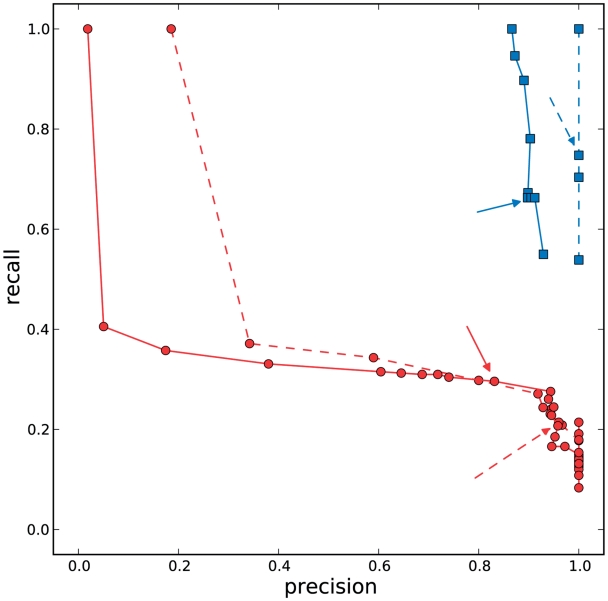
Figure 2.
Precision–recall analysis. We considered the haplotypes inferred in all windows by the clustering algorithm and by the cut-off method based on the minimum number of reads supporting the variant. Red circles represent precision and recall for a set of threshold values chosen in the cut-off method (values from 1 to the number of reads in the most-supported haplotype), red arrows annotate points for a cut-off equal to 50. We performed a similar analysis on the output of the clustering algorithm, considering haplotypes whose confidence value (posterior probability) was greater or equal than a given threshold. Blue squares represent threshold values from 0.01 to 1, with blue arrows annotating the values obtained when the threshold is 0.9. Dashed lines and arrows are used for points obtained in the non-PCR-ampified sample, solid lines and arrows for points in the PCR-amplified one. In the non-PCR-amplified sample, we have a perfect precision (no false positives), and very good results for the recall. In the PCR-amplified sample, some false positives are found. In both cases, the performance of the clustering method is superior to the cut-off method. Results for individual windows can be found in Supplementary Data.