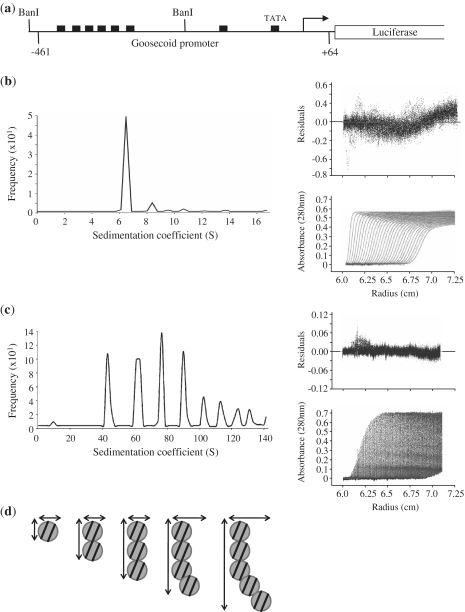
Figure 2.
The formation of large PRH–DNA complexes. (a) A schematic of the Goosecoid reporter construct used in this study. Goosecoid promoter sequences from –461 to +64 relative to the transcription start point (bent arrow) are located upstream of the luciferase gene. The filled rectangles represent core PRH binding sites and the Goosecoid TATA box sequence. The AUC experiments described below were performed with a 267-bp DNA fragment (96 nM) produced by digestion with BanI. (b) Sedimentation velocity experiments were performed with the BanI fragment shown in (a). Sedimentation was monitored as a function of centrifugal radius using the absorbance at 260 nm (DNA). A single boundary is observed corresponding to the free DNA (bottom right panel) with a sedimentation coefficient of around 7 S (main panel). (c) In the presence of PRH several boundaries are observed corresponding to free and bound DNA (bottom right panel) with sedimentation coefficients of 41 S, 65 S, 86 S, 104 S and 120 S. (d) A model for the formation of PRH–DNA complexes based on the data shown in Table 1. Circles represent PRH double octamers. The lines represent 267-bp DNA fragments.