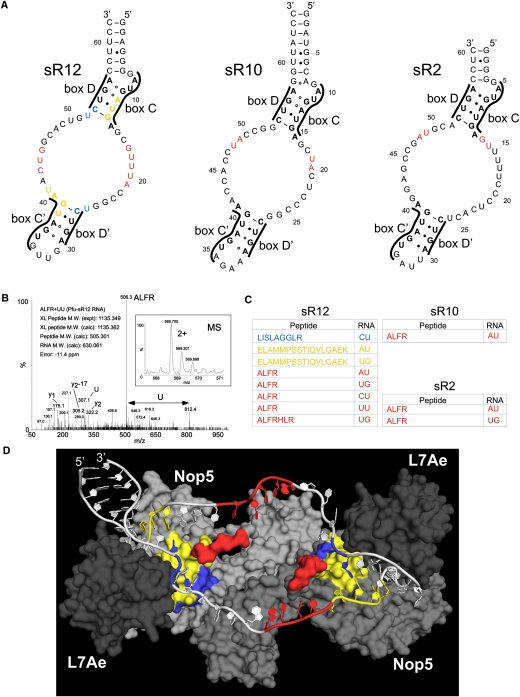
FIGURE 2.
The AFLR motif contacts the guide/spacer regions of the box C/D sRNA. (A) Secondary structures of the P. furiosus sRNAs. The box motifs are indicated. The cross-link sites are colored according to panel C. (B) MS and MSMS spectra of a peptide–RNA cross-link derived from in vitro assembled RNP with sR12 RNA. The figure shows the intact mass and the charge state of the precursor that was selected for MSMS (small box) and the corresponding MSMS spectra that identified the cross-linked peptide and RNA moiety. Y-type fragment ions derived from the cross-linked peptide under CID (collision induced decay) are assigned as well as the m/z values for the nucleotide U in the lower m/z regime. (Arrow) The mass difference that corresponds exactly to a U. The experimentally determined precursor (cross-linked peptide–RNA oligonucleotide), the calculated peptide and RNA mass, and the mass deviation (error) in parts per million (ppm) are listed within the figure. (C) Cross-linked peptides and RNAs. In vitro assembled sRNPs were denatured and digested with trypsin and RNAses. RNA–peptide conjugates were purified and then characterized by ESI-mass spectrometric analysis. The colors correspond to the RNA and protein sites in panels A and D. (D) Structural model of sR12 box C/D sRNP showing RNA and protein contact sites. The structure of the Nop5, L7Ae, sRNA complex published by Ye et al. (2009) was modified to include the sR12 sRNA. Alanines were also used to join the section of the protuberance missing from the crystal structure. A surface view is shown for the proteins with Nop5 (gray and light gray) and L7Ae (dark gray) with the cross-linked peptides colored as in panel C. For clarity, we have not included fibrillarin in the image. The RNA is shown in white, with the cross-linked nucleotides colored as in panels A and C.