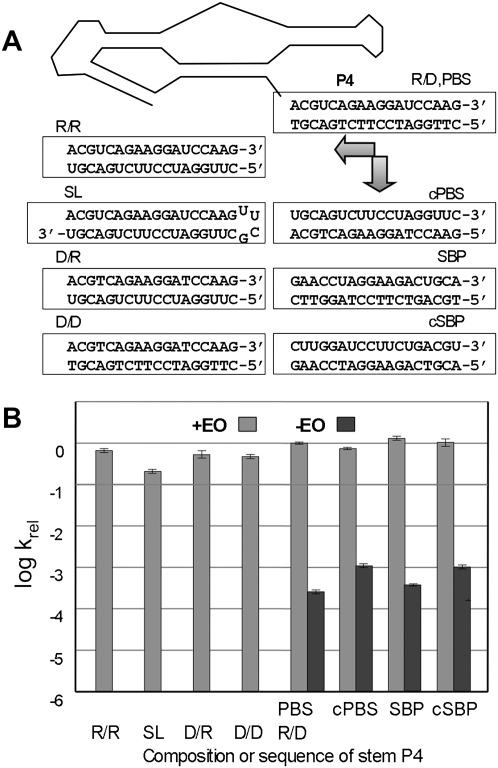
FIGURE 4.
Effect of P4 helix composition on ribozyme activity. (A) P4 variants are shown below a schematic of Kin.46. “SL” is a stem–loop construct in which the ribozyme 3′ end folds back on itself to form the activating helix. Essentially identical data were obtained for an SL construct in which the UUCG loop was replaced with a slightly less stable GAAA tetraloop (Supplemental Table S1; data not shown). (B) Relative apparent initial rate constants for P4 variants shown in A. Apparent binding affinity for donor substrates is not adversely affected by altering the sequence of the 3′ tail. Km values for all four tail variants are within experimental error of each other, both in the presence or absence of EO (0.5–1.6 ± 0.4 μM) (Supplemental Table S1), and all are well below the substrate concentration used in the Eyring analysis (10 μM).