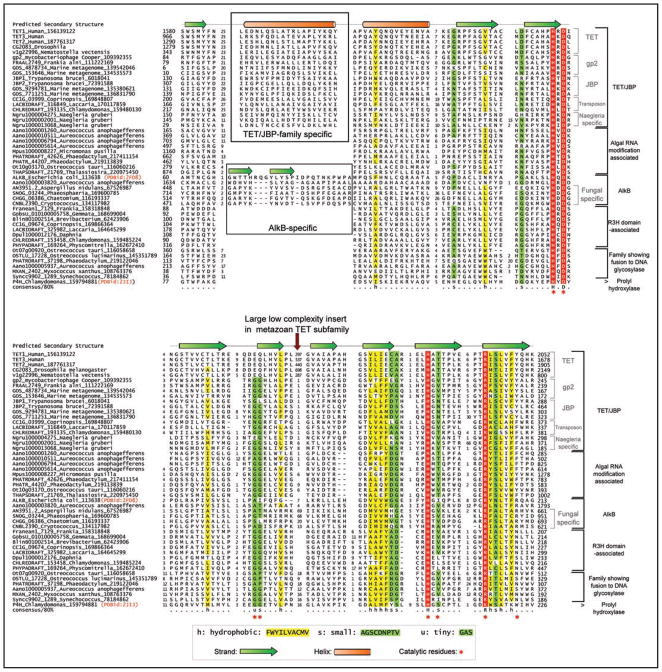
Figure 1.
Multiple alignment of selected examples of the newly predicted families of the nucleic-acid-modifying 2-oxoglutarate- and iron(II)-dependent dioxygenase superfamily. Protein sequences are represented by their gene names, species names and GenBank index numbers (where available). Temporary gene names were assigned for predicted proteins from Naegleria, Aureococcus, Daphnia and Micromonas. The full length protein sequences from these are available in the Supplementary material. The coloring scheme and consensus abbreviations are shown in the key. Family names are shown to the right of the alignment. The distinct inserts of the TET/JBP and the AlkB families are shown within boxes. The key conserved residues defining the 2OGFeDO protein have been marked below the alignment. The consensus secondary structure derived from crystal structures of characterized members of the superfamily is shown above.