Abstract
Objective. Combined cisplatin (CDDP) and radiotherapy is increasingly being used to treat advanced head and neck cancers. As both CDDP and radiation can cause hearing loss, it is important to have a better understanding of the cellular and molecular ototoxic mechanisms involved in combined therapy. Procedure. The effects of CDDP, radiation, and combined CDDP-radiation on the OC-k3 cochlear cell line were studied using MTS assay, flow cytometry, Western blotting, and microarray analysis. Results. Compared to using CDDP or radiation alone, its combined use resulted in enhanced apoptotic cell death and apoptotic-related gene expression, including that of FAS. Phosphorylation of p53 at Ser15 (a marker for p53 pathway activation in response to DNA damage) was observed after treatment with either CDDP or radiation. However, posttreatment activation of p53 occurred earlier in radiation than in CDDP which corresponded to the timings of MDM2 and TP53INP1 expression. Conclusion. Enhanced apoptotic-related gene expressions leading to increased apoptotic cell deaths could explain the synergistic ototoxicity seen clinically in combined CDDP-radiation therapy. CDDP and radiation led to differential temporal activation of p53 which suggests that their activation is the result of different upstream processes. These have implications in future antiapoptotic treatments for ototoxicity.
1. Introduction
Combined chemoradiotherapy is increasingly being used to treat advanced head and neck cancers. During radiotherapy, the ear structures are often included in the radiation fields and it is generally accepted that radiation-induced sensorineural hearing loss can result. Cisplatin (CDDP), widely used as an effective antineoplastic drug for these cancers, is also known to cause ototoxicity. In a randomized blinded study, it was demonstrated that patients who had received radiotherapy and concurrent/adjuvant chemotherapy using CDDP experienced greater sensorineural hearing loss compared with patients treated with radiotherapy alone [1]. This was especially so in the high-frequency sounds of the speech range, resulting in significant hearing disability.
In recent years, immortalized cell lines derived from the mouse organ of Corti had been developed and characterized [2]. For example, the OC-k3 cell line was derived from the organ of Corti of the transgenic mouse. It encoded the large T antigen of the SV40 (simian virus 40), a thermolabile viral protein which drove the cells to proliferate indefinitely at 33°C and in the presence of gamma interferon [3]. This cell line expressed the neuro-epithelial precursor cell marker nestin and the inner ear cell marker OCP2, but did not exhibit markers for glial or neuronal cells. In addition, OC-k3 cells expressed specific auditory sensory cell markers (myosin VIIa and the acetylcholine receptor alpha-9) and the supporting cell marker connexin 26. This and other similar cell lines had been regarded as good models to study the mechanisms of cell fate in the organ of Corti of the cochlea [4].
P53 had been found to play an important role in apoptotic cell death associated with ototoxicity. In a CDDP-induced apoptosis experiment using cochlear organotypic cultures prepared from rats at postnatal days 3-4, significant upregulation of phospho-p53 serine 15 expression was found and apoptosis was suppressed by pifithrin-α, a p53 inhibitor [5]. Other studies have shown that the deletion of the p53 gene protects sensory hair cells from CDDP-induced cell death, caspase-2 activation, and cytochrome c translocation [6]. In radiation-induced ototoxicity, it was found that p53 together with reactive oxidative species (ROS) played an important role in cochlear cell apoptosis [7].
In the combined use of CDDP and radiation, the cellular and molecular mechanisms leading to ototoxicity had not been studied. It is important to have a better understanding of these mechanisms as effective preventive strategies directed at the relevant pathways can potentially be developed. The present study found that although p53 played a role in both CDDP and radiation-induced cochlear cell apoptosis, p53 was activated at different time points after each treatment which corresponded to the time MDM2 and TP53INP1 were expressed. Additional apoptotic-related genes that were not expressed when CDDP or radiation was used alone were expressed when used in combination. This included FAS, an important element involved in the extrinsic apoptotic pathway.
2. Materials and Methods
2.1. Cell Culture
The immortalized OC-k3 cell line derived from the organ of Corti of the transgenic mice (Immortamouse H-2Kb-tsA58, Charles Rivers Laboratories, Wilmington, MA) was used. The cell line was cultured in high-glucose Dulbecco's Eagle's medium (DMEM, Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY), 1% penicillin-streptomycin (P/S, Gibco, Grand Island, NY), and 50 U/ml gamma-interferon (mouse recombinant, Sigma-Aldrich, St. Louis, MO) and maintained at 33°C with 10% CO2. To study the impact of chemoradiation treatment, OC-k3 cells were exposed to 5 Gy of gamma irradiation alone, 0.5 μg/ml of cisplatin alone, or 5 Gy of gamma irradiation in the presence of 0.5 μg/ml cisplatin (Pfizer, Bentley, WA).
2.2. Cell Viability Assay
The OC-k3 cells were seeded in 96-well plates at densities of 5 × 103 cells/well in 200 μl complete medium after being exposed to chemo-irradiation treatment. Cell viability was determined using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega Corp., Madison, WI) containing tetrazolium compound 3-[4,5-dimethylthiazol-2-yl]-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) at 3 h, 24 h, 48 h, and 72 h after chemo-irradiation. This test was based on the bioreduction of MTS compound into a soluble and colored formazan product by NADPH or NADH, which is produced by dehydrogenase enzymes in metabolically active cells. Twenty microliters of MTS were added to each well, incubated at 33°C for 3 h, and then the absorbance was recorded at 490 nm with a microplate spectrophotometer (Benchmark Plus, Bio-Rad Laboratories, Hercules, CA).
2.3. Cell Death Analysis
The cells were collected at each time point post CDDP-radiation treatment, fixed in 75% ethanol and stored at 4°C. Upon analysis, the cells were washed with PBS and incubated with 100 μg/ml propidium iodide (PI) containing 0.1% Triton X-100 and 500 μg/ml RNase A in 50 μl PBS for 30 mins in darkness at 4°C. The DNA contents of cells were analyzed using the flow cytometer CyAnTM ADP Analyser (Beckman Coulter, Fullerton, CA). The magnitudes of the sub-G1 fractions were determined using the Summit 4.3 software (Beckman Coulter, Fullerton, CA). DNA fragmentation resulting from apoptotic cell death would manifest in the sub-G1 fraction.
2.4. Western Blot Analysis
Protein extraction was done by incubating the cells at 4°C for 30 minutes in lysis buffer containing 150 mM NaCl, 10 mM Tris-HCl pH 7.4, 2 mM EDTA, 0.5 mM EGTA, 1 mM sodium orthovanadate, 0.1% sodium deoxycholate, 0.5% NP-40, and 1% Triton X-100 supplemented with 1x complete protease inhibitor mixture (Roche, Basel, Switzerland). Equal amounts of protein samples were denatured separated by 10% SDS-PAGE and transferred onto nitrocellulose membrane by iBlot dry blotting system (Invitrogen, Carlsbad, CA). The membrane was blocked with 5% nonfat milk in PBS with 0.1% Tween-20 (PBST) for 1 h, followed by an overnight incubation of primary antibodies in 5% BSA/PBST at 4°C. Primary antibodies included anti-p53 pAb (NCL-p53-CM5p, Novocastra), anti-phospho-p53 (ser-15) pAb, anti-phospho-c-jun (ser-73) pAb, anti-c-jun (60A8) mAb (Cell Signaling Technology, Inc.), and anti-beta-actin mAb (Sigma-Aldrich, St. Louise, MO). After washing the membrane extensively, incubation with horseradish peroxidase-conjugated antirabbit or antimouse secondary antibody (Cell Signaling Technology, Inc.) was done for 1 h at room temperature. After washing, the membrane was incubated in Immobilon Western chemiluminescent HRP substrate (Millipore, Billerica, MA), and the chemiluminescence signals were detected using UVIchemi (UVItec, Cambridge, UK), a dedicated chemiluminescence documentation system. For reprobing with a new primary antibody, the membrane was stripped in Re-Blot plus strong solution Western blot stripping buffer (Chemicon, Temecula, CA) at room temperature for 30 minutes and rinsed 3 times with PBST for 10 minutes each time.
2.5. Microarray Analysis
The global changes of gene expression were analyzed at 3 h, 24 h, and 72 h after chemoirradiation, on the GeneChip Mouse Genome 430A 2.0 Array (Affymetrix, Santa Clara, CA). Biological duplicates of experiments were performed. Briefly, RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA) followed by generation of double-stranded cDNA. These were used as templates for synthesis of biotin-labeled cRNA, using the GeneChip IVT labeling kit in accordance with the manufacturer's instructions. The biotinylated cRNA was purified using RNeasy Mini kit (Qiagen, Hilden, Germany) and fragmented before reconstitution in a hybridization cocktail mixture containing eukaryotic hybridization control. The hybridization was performed at 45°C for 16 h in a rotisserie oven set at 60 rpm. Upon completion, the arrays were then loaded onto an Affymetrix Fluidic station, washed according to the standard Affymetrix EukGE-WS2v5 protocol and stained with streptavidin-phycoerythrin (SAPE) solution. After washing and staining, the arrays were scanned with the Gene Array scanner (Affymetrix, Santa Clara, CA). Hybridization intensity data detected by the scanner were automatically acquired and processed by the Affymetrix GeneChip Operating Software (GCOS, Affymetrix, Santa Clara, CA). The average intensity for all the genes was normalized to 100. The statistical algorithms implemented in GCOS software were used for analysis. In a comparison expression analysis, each probe pair on the experimental array was compared to the corresponding probe pair on the baseline array (control). This generated an associated “change” (increased, no change, or decreased) to determine the relative expression of transcripts. To have an overview of gene expression profiles, probe sets showing chemoradiation-induced increased or decreased expressions in both duplicated experiments were retrieved. The differentially expressed genes of chemoradiation treatment were submitted for biological functional analysis using Ingenuity Pathway Analysis (IPA) tools (Ingenuity Systems, http://www.ingenuity.com).
3. Results
3.1. Combined CDDP-Radiation Treatment Reduced Cell Viability More than CDDP or Radiation Treatment Alone
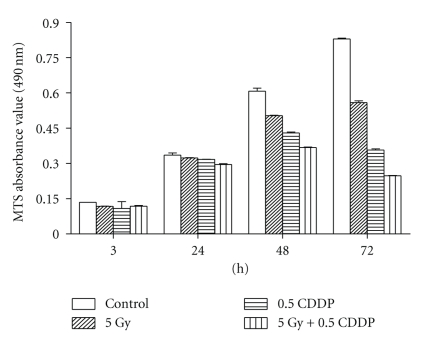
Cell viability analysis by MTS assay at different time points revealed that although CPPD and radiation each exerted a negative effect on cell viability, treatment when combined appeared to have a greater effect. These effects were observed at 48 hrs after treatment and became even more marked at 72 hrs after treatment (Figure 1).
Figure 1.
Cell viability analysis by MTS assay at different time points (3 h, 24 h, 48 h, and 72 h) after treatment with 5 Gy gamma radiation and 0.5 μg/ml cisplatin (CDDP). After co-treatment with radiation and CDDP, cell viability was significantly reduced at 72 h. The data shown are the most representative of 3 separate experiments.
3.2. Apoptosis Occurred Predominantly at 72 h after Combined CDDP-Radiation Treatment
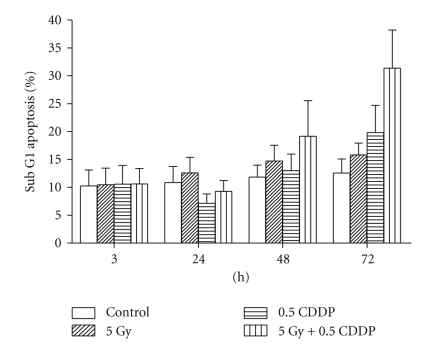
At 72 hrs after treatment, combined CDDP-radiation led to a greater increase in the sub-G1 phase as compared to using CDDP and radiation alone (Figure 2). As pointed out previously, DNA fragmentation resulting from apoptotic cell death manifests in the sub-G1 fraction.
Figure 2.
Flow cytometric subG1 phase as determined by PI staining at different time points (3 h, 24 h, 48 h and 72 h) after exposure to 5 Gy of gamma radiation and 0.5 μg/ml of cisplatin (CDDP). Co-treatment with radiation and CDDP resulted in a significant increase in subG1 phase at 72 h. The data shown are the mean + SD of 4 independent experiments.
3.3. Apoptosis-Related Gene Expressions were Enhanced by Combined CDDP-Radiation Treatment
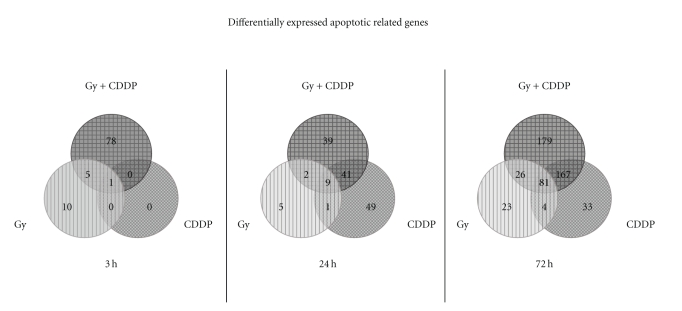
On analyzing the results of molecular and cellular functions under the biological functions of IPA, it was found that among the 3925 probe set IDs which were differentially expressed in at least one treatment, 942 represented 623 unique genes associated with apoptosis (see Table 1). Their distribution at each time point for the different treatment regimes is summarized in Venn diagrams (Figure 3). A subset focusing on the genes, which had a direct upstream or downstream relationship with p53, is shown in Table 2. Combined CDDP-radiation treatment resulted in an increase in the number of gene expressions which was more than merely a summation of the number of expressions resulting from individual treatments (Figure 3, Table 2). At 72 hrs after treatment, 40 out of the 163 genes listed (24.5%) were expressed in combined CDDP-radiation treatment, but not when CDDP or radiation was used alone (Table 2). Among these 40 genes was FAS, an important element of the extrinsic apoptotic pathway.
Table 1.
Differentially expressed apoptosis-related genes in each treatment group [irradiation (Gy), cisplatin (CDDP), or combination of both (Gy + CDDP)] when compared to nontreated control cells at 3 h, 24 h, and 72 h after treatment
| 3 h | 24 h | 72 h | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Symbol | Probe set ID | Gy | CDDP | Gy + CDDP | Gy | CDDP | Gy + CDDP | Gy | CDDP | Gy + CDDP |
| AAK1 | 1420025_s_at, 1434935_at | I | I | |||||||
| AARS | 1423685_at | D | ||||||||
| ABCB1B | 1418872_at | I | I | |||||||
| ABCC1 | 1452233_at | I | ||||||||
| ABCG2 | 1422906_at | I | ||||||||
| ABL2 | 1455495_at | I | ||||||||
| ACSL4 | 1451828_a_at | I | ||||||||
| ACTN4 | 1423449_a_at | I | ||||||||
| ACVR1 | 1448460_at | D | ||||||||
| ADAMTSL4 | 1451932_a_at | I | ||||||||
| ADM | 1416077_at, 1447839_x_at | D | D | I | I | |||||
| AES | 1420619_a_at | D | ||||||||
| AFP | 1416645_a_at | I | ||||||||
| AHR | 1422631_at | I | ||||||||
| AIMP1 | 1416486_at | D | ||||||||
| AKAP12 | 1419706_a_at | I | I | |||||||
| AKT1S1 | 1428158_at, 1452684_at | D | ||||||||
| ALDH1A1 | 1416468_at | I | I | |||||||
| ALDH1A2 | 1422789_at | D | ||||||||
| ALDOA | 1416921_x_at, 1433604_x_at, 1434799_x_at, 1439375_x_at | D | D | I | I | I | ||||
| ANKRD1 | 1420991_at, 1420992_at | I | I | I | ||||||
| ANLN | 1433543_at, 1439648_at | I | I | |||||||
| ANP32A | 1421918_at | D | ||||||||
| ANXA1 | 1448213_at | I | I | I | ||||||
| ANXA7 | 1416138_at | I | I | |||||||
| AP2A2 | 1452490_a_at | I | ||||||||
| APBB2 | 1452342_at | I | ||||||||
| APEX1 | 1416135_at, 1437715_x_at, 1456079_x_at | D | D | |||||||
| APOE | 1432466_a_at | I | I | I | ||||||
| APPL1 | 1455159_at | I | ||||||||
| ARNT2 | 1434028_at | I | ||||||||
| ASAH2 | 1450726_at | I | I | |||||||
| ATF3 | 1449363_at | D | D | D | ||||||
| ATF5 | 1425927_a_at | D | D | |||||||
| ATG12 | 1451746_a_at | D | D | |||||||
| ATG5 | 1418235_at | I | ||||||||
| ATM | 1421205_at, 1428830_at | I | I | |||||||
| ATN1 | 1421149_a_at | I | ||||||||
| ATP1A1 | 1451071_a_at | D | ||||||||
| ATP7A | 1436921_at | I | ||||||||
| ATXN2 | 1460653_at | D | ||||||||
| AURKA | 1424511_at | D | I | I | I | |||||
| AXL | 1423586_at | D | I | I | ||||||
| BAG3 | 1422452_at | D | ||||||||
| BANF1 | 1421081_a_at, 1421082_s_at, 1421083_x_at | D | D | |||||||
| BCAR1 | 1439388_s_at, 1450622_at | D | I | I | ||||||
| BCL3 | 1418133_at | I | I | |||||||
| BCLAF1 | 1428844_a_at, 1428845_at, 1436023_at, 1438089_a_at | I | I | |||||||
| BDNF | 1422168_a_at | I | ||||||||
| BECN1 | 1455880_s_at, 1460320_at | I | I | |||||||
| BGN | 1437889_x_at, 1448323_a_at | D | D | I | ||||||
| BHLHE40 | 1418025_at | D | D | I | ||||||
| BID | 1417045_at, 1448560_at | D | I | I | ||||||
| BIRC3 | 1421392_a_at | I | ||||||||
| BIRC5 | 1424278_a_at | I | I | |||||||
| BLM | 1448953_at | I | ||||||||
| BNIP2 | 1453993_a_at | D | ||||||||
| BNIP3 | 1422470_at | D | ||||||||
| BPTF | 1427310_at, 1456615_a_at | I | ||||||||
| BRAF | 1435434_at | I | ||||||||
| BRCA1 | 1424629_at, 1424630_a_at, 1451417_at | I | ||||||||
| BRD2 | 1423502_at, 1437210_a_at | D | D | D | ||||||
| BRE | 1426312_at, 1426313_at | D | D | D | ||||||
| BTG1 | 1426083_a_at | D | ||||||||
| BTG2 | 1416250_at, 1448272_at | I | I | |||||||
| BUB1 | 1424046_at | I | I | I | I | |||||
| BUB1B | 1416961_at, 1447363_s_at | I | I | I | ||||||
| C11ORF82 | 1429734_at | I | I | I | ||||||
| C1QBP | 1455821_x_at | D | ||||||||
| C3 | 1423954_at | I | I | |||||||
| CACNA1A | 1450510_a_at | I | ||||||||
| CACNA1C | 1421297_a_at | D | D | |||||||
| CASP12 | 1449297_at | D | ||||||||
| CASP2 | 1448165_at | D | ||||||||
| CASP3 | 1426165_a_at, 1449839_at | I | ||||||||
| CASP6 | 1415995_at | I | I | |||||||
| CASP7 | 1426062_a_at, 1448659_at | D | D | |||||||
| CASP9 | 1426125_a_at | D | ||||||||
| CAST | 1426098_a_at, 1435972_at, 1451413_at | I | I | I | I | I | ||||
| CAT | 1416429_a_at | I | ||||||||
| CAV1 | 1449145_a_at | D | I | I | I | |||||
| CBX5 | 1421933_at, 1450416_at | D | D | |||||||
| CCAR1 | 1436156_at, 1436157_at | I | I | |||||||
| CCL13 | 1420380_at | I | I | I | I | |||||
| CCL5 | 1418126_at | I | I | I | I | |||||
| CCL9 | 1417936_at, 1448898_at | I | I | |||||||
| CCNA2 | 1417910_at, 1417911_at | I | I | |||||||
| CCNB1 | 1416076_at, 1419943_s_at, 1448205_at, 1449675_at | D | I | I | I | |||||
| CCND1 | 1417419_at, 1417420_at, 1448698_at | D | I | I | I | |||||
| CCND3 | 1415907_at | I | I | |||||||
| CCNG1 | 1420827_a_at, 1450016_at, 1450017_at | I | I | I | I | I | I | I | I | |
| CD14 | 1417268_at | I | ||||||||
| CD24 | 1416034_at, 1437502_x_at, 1448182_a_at | D | I | |||||||
| CD274 | 1419714_at | I | D | |||||||
| CD2AP | 1420907_at | I | ||||||||
| CD44 | 1423760_at, 1434376_at, 1452483_a_at | I | I | |||||||
| CD47 | 1419554_at, 1428187_at, 1449507_a_at | D | D | |||||||
| CD80 | 1432826_a_at | I | ||||||||
| CD9 | 1416066_at | D | I | I | I | |||||
| CDC20 | 1416664_at, 1439377_x_at | I | I | I | ||||||
| CDC25B | 1421963_a_at | I | I | |||||||
| CDC25C | 1422252_a_at, 1456077_x_at | I | I | |||||||
| CDC2L2 | 1418841_s_at | D | ||||||||
| CDC37 | 1416819_at | I | I | |||||||
| CDC42EP3 | 1422642_at, 1450700_at | I | I | I | ||||||
| CDC45L | 1416575_at | D | D | |||||||
| CDC6 | 1417019_a_at | D | ||||||||
| CDCA2 | 1437251_at, 1455983_at | I | I | |||||||
| CDH2 | 1418815_at | I | ||||||||
| CDK4 | 1422439_a_at, 1422440_at, 1422441_x_at | D | D | D | ||||||
| CDK8 | 1460389_at | I | ||||||||
| CDKN1A | 1421679_a_at, 1424638_at | I | I | I | I | I | I | I | I | |
| CDKN1B | 1434045_at | D | D | |||||||
| CDKN2A | 1450140_a_at | D | ||||||||
| CDKN2C | 1416868_at | D | ||||||||
| CEBPB | 1427844_a_at | D | D | |||||||
| CEBPD | 1423233_at | D | I | I | I | |||||
| CENPF | 1427161_at | I | ||||||||
| CFLAR | 1424996_at | I | ||||||||
| CHEK1 | 1439208_at | D | ||||||||
| CKAP2 | 1434748_at | I | I | I | I | |||||
| CLCF1 | 1437270_a_at, 1450262_at | D | I | |||||||
| CLU | 1418626_a_at, 1437458_x_at, 1437689_x_at, 1454849_x_at | I | I | I | ||||||
| CNN2 | 1450981_at | I | I | |||||||
| CNP | 1418980_a_at, 1437341_x_at | I | I | |||||||
| CNTF | 1426327_s_at | D | ||||||||
| COPS5 | 1460171_at | D | ||||||||
| CR1 | 1422563_at | D | ||||||||
| CREB3L1 | 1419295_at | I | I | |||||||
| CRK | 1416201_at, 1448248_at | D | I | I | I | |||||
| CROP | 1424802_a_at, 1451485_at | I | I | D | ||||||
| CRYAB | 1416455_a_at, 1434369_a_at | I | I | I | ||||||
| CSF1 | 1425154_a_at, 1425155_x_at, 1448914_a_at, 1460220_a_at | D | I | I | ||||||
| CSF2 | 1427429_at | I | ||||||||
| CSNK2A1 | 1419034_at, 1419035_s_at, 1419036_at, 1419038_a_at | D | D | |||||||
| CST3 | 1426195_a_at | I | ||||||||
| CTCF | 1418330_at, 1449042_at | D | D | |||||||
| CTGF | 1416953_at | I | I | I | ||||||
| CTNNA1 | 1437807_x_at, 1448149_at | I | ||||||||
| CTSB | 1417490_at, 1417491_at, 1417492_at | I | I | |||||||
| CTSD | 1448118_a_at | I | I | |||||||
| CTTN | 1421313_s_at, 1421315_s_at, 1423917_a_at, 1433908_a_at | I | I | |||||||
| CUL3 | 1434717_at | D | ||||||||
| CUL5 | 1428287_at | I | ||||||||
| CX3CL1 | 1415803_at | I | ||||||||
| CXCL12 | 1417574_at, 1448823_at | D | D | |||||||
| CXCL2 | 1419209_at, 1441855_x_at, 1457644_s_at | D | I | I | I | |||||
| CXCR7 | 1417625_s_at | D | D | I | I | |||||
| CYB5A | 1416727_a_at | I | ||||||||
| CYB5R3 | 1422185_a_at, 1422186_s_at, 1425329_a_at | I | I | |||||||
| CYBA | 1454268_a_at | I | ||||||||
| CYLD | 1429617_at | I | ||||||||
| CYR61 | 1416039_x_at, 1438133_a_at, 1442340_x_at, 1457823_at | I | I | I | ||||||
| DAB2 | 1420498_a_at, 1423805_at, 1429693_at | I | I | |||||||
| DAP | 1423790_at, 1451112_s_at | D | D | I | I | I | ||||
| DAXX | 1419026_at | D | I | |||||||
| DCN | 1449368_at | I | ||||||||
| DDIT3 | 1417516_at | D | ||||||||
| DDIT4 | 1428306_at | I | I | I | ||||||
| DDR1 | 1415797_at, 1415798_at, 1456226_x_at | D | I | I | ||||||
| DDX5 | 1419653_a_at | I | ||||||||
| DDX58 | 1436562_at, 1456890_at | D | I | D | ||||||
| DHCR24 | 1451895_a_at | I | I | |||||||
| DKK3 | 1417312_at, 1448669_at | D | D | I | I | I | ||||
| DLC1 | 1436173_at, 1460602_at | I | ||||||||
| DLX2 | 1448877_at | D | ||||||||
| DNAJC15 | 1416910_at | D | ||||||||
| DNM1L | 1428086_at, 1452638_s_at | I | ||||||||
| DTYMK | 1438096_a_at | I | ||||||||
| DUSP14 | 1431422_a_at | I | I | |||||||
| DUSP22 | 1448985_at | I | ||||||||
| DUSP4 | 1428834_at | D | ||||||||
| DUSP6 | 1415834_at | I | ||||||||
| DUT | 1419270_a_at | D | ||||||||
| E2F1 | 1417878_at | D | ||||||||
| ECOP | 1451127_at | D | ||||||||
| EDA2R | 1440085_at | I | I | I | I | I | ||||
| EEF1D | 1439439_x_at, 1449506_a_at | D | D | |||||||
| EGR1 | 1417065_at | I | I | I | ||||||
| EHD4 | 1449852_a_at | I | ||||||||
| EIF2AK2 | 1422006_at, 1440866_at | I | I | I | ||||||
| EIF4E | 1450908_at | D | ||||||||
| EIF5A | 1437859_x_at | D | ||||||||
| ELAVL1 | 1452858_at | D | ||||||||
| EMILIN2 | 1435264_at | I | I | |||||||
| EMP1 | 1416529_at | I | I | I | ||||||
| EMP3 | 1417104_at | I | I | I | ||||||
| ENO1 | 1419022_a_at, 1419023_x_at | I | ||||||||
| EPHA2 | 1421151_a_at | I | ||||||||
| EPHX1 | 1422438_at | I | I | I | ||||||
| ERCC3 | 1448497_at | I | ||||||||
| ERCC5 | 1450935_at | I | ||||||||
| ESPL1 | 1433862_at | I | ||||||||
| ETS1 | 1422027_a_at, 1426725_s_at, 1452163_at | D | I | |||||||
| ETS2 | 1416268_at | D | ||||||||
| EVI1 | 1438325_at | D | D | |||||||
| EWSR1 | 1417238_at | I | I | |||||||
| EXOC2 | 1428470_at | I | ||||||||
| EZR | 1450850_at | I | I | |||||||
| F2R | 1437308_s_at, 1450852_s_at | I | ||||||||
| F3 | 1417408_at | D | ||||||||
| FAS | 1460251_at | I | ||||||||
| FASN | 1423828_at | I | ||||||||
| FBL | 1416684_at, 1416685_s_at | D | D | |||||||
| FBN1 | 1425896_a_at, 1438870_at, 1460208_at | I | I | |||||||
| FDFT1 | 1438322_x_at, 1448130_at | D | I | I | ||||||
| FEN1 | 1421731_a_at, 1436454_x_at | D | D | D | ||||||
| FGF7 | 1422243_at, 1438405_at | I | I | I | ||||||
| FGFR1 | 1424050_s_at | D | I | I | ||||||
| FHL2 | 1419184_a_at | D | I | I | ||||||
| FKBP1B | 1449429_at | I | I | |||||||
| FLT3LG | 1422115_a_at | I | ||||||||
| FN1 | 1437218_at | I | ||||||||
| FOS | 1423100_at | I | I | |||||||
| FOSL1 | 1417487_at, 1417488_at | I | I | I | ||||||
| FOXM1 | 1417748_x_at, 1448833_at, 1448834_at, 1453107_s_at | I | I | |||||||
| FOXO1 | 1416982_at | D | ||||||||
| FOXP1 | 1421141_a_at, 1421142_s_at, 1435222_at | D | ||||||||
| FST | 1421365_at, 1434458_at | I | I | I | I | |||||
| FSTL1 | 1416221_at, 1448259_at | I | I | I | ||||||
| FTH1 | 1427021_s_at | I | ||||||||
| FUBP1 | 1433482_a_at, 1433640_at | I | D | D | ||||||
| FUS | 1451285_at, 1455831_at | I | D | |||||||
| FXN | 1427282_a_at | D | ||||||||
| FXR1 | 1417598_a_at, 1442059_at, 1452247_at | I | ||||||||
| FYN | 1448765_at | I | ||||||||
| G2E3 | 1434699_at, 1455355_at | I | I | |||||||
| G6PD | 1448354_at | I | I | |||||||
| GABPA | 1450665_at | D | ||||||||
| GADD45A | 1449519_at | D | D | D | ||||||
| GAS1 | 1416855_at, 1448494_at | D | D | D | D | D | D | |||
| GATAD2A | 1423992_at, 1451197_s_at, 1451198_at, 1455505_at | D | I | |||||||
| GDF15 | 1418949_at | I | I | |||||||
| GDNF | 1419080_at | I | ||||||||
| GFRA1 | 1450440_at | I | I | |||||||
| GHR | 1417962_s_at, 1451501_a_at | I | I | |||||||
| GJA1 | 1415800_at, 1415801_at, 1437992_x_at, 1437992_x_at, 1438650_x_at, 1438945_x_at, 1438973_x_at | D | D | I | I | |||||
| GLIPR1 | 1424927_at | I | I | I | ||||||
| GLRX | 1416592_at, 1416593_at | I | I | |||||||
| GNA12 | 1421026_at, 1450097_s_at | D | ||||||||
| GNA13 | 1422556_at, 1433749_at, 1450656_at, 1453470_a_at, 1460317_s_at | D | D | |||||||
| GNPNAT1 | 1423158_at | D | ||||||||
| GPI | 1420997_a_at, 1434814_x_at, 1450081_x_at | I | ||||||||
| GPX1 | 1460671_at | I | ||||||||
| GRN | 1448148_at | I | ||||||||
| GSK3B | 1437001_at, 1451020_at, 1454958_at | D | D | |||||||
| GSN | 1415812_at, 1436991_x_at, 1437171_x_at, 1456312_x_at | I | I | |||||||
| GSPT1 | 1426736_at, 1452168_x_at | D | ||||||||
| GSTM1 | 1416411_at | I | ||||||||
| GSTM5 | 1448330_at | D | ||||||||
| HBEGF | 1418349_at | D | ||||||||
| HELLS | 1417541_at | I | ||||||||
| HIP1 | 1434557_at | D | ||||||||
| HIPK1 | 1424540_at | D | ||||||||
| HIST1H1C | 1416101_a_at, 1436994_a_at | D | D | I | I | |||||
| HK1 | 1420901_a_at | I | I | |||||||
| HK2 | 1422612_at | I | ||||||||
| HMGA1 | 1416184_s_at | I | I | I | ||||||
| HMGA2 | 1422851_at, 1450780_s_at, 1450781_at | I | I | I | ||||||
| HMGB1L1 | 1425048_a_at, 1435324_x_at, 1439463_x_at, 1448235_s_at | D | D | |||||||
| HMGN1 | 1455897_x_at | D | ||||||||
| HMMR | 1425815_a_at, 1427541_x_at, 1450156_a_at, 1450157_a_at | I | I | I | ||||||
| HMOX1 | 1448239_at | D | ||||||||
| HNRNPA1 | 1423531_a_at, 1430019_a_at, 1430020_x_at | D | D | D | ||||||
| HOXA7 | 1449499_at | D | ||||||||
| HSH2D | 1442130_at | I | I | |||||||
| HSP90AA1 | 1426645_at, 1437497_a_at, 1438902_a_at | I | I | I | ||||||
| HSP90AB1 | 1416364_at, 1416365_at | I | ||||||||
| HSPA1B | 1427127_x_at | D | ||||||||
| HSPA5 | 1416064_a_at, 1427464_s_at, 1447824_x_at | D | D | |||||||
| HSPB1 | 1422943_a_at, 1425964_x_at | D | I | I | I | |||||
| HSPB8 | 1417014_at | D | ||||||||
| HTATIP2 | 1451814_a_at | I | ||||||||
| HUWE1 | 1415703_at | D | ||||||||
| ID1 | 1425895_a_at | I | ||||||||
| ID2 | 1422537_a_at | D | ||||||||
| IER3 | 1419647_a_at | I | I | I | ||||||
| IFI16 | 1419603_at, 1452349_x_at | I | D | |||||||
| IFI202B | 1421551_s_at, 1457666_s_at | I | I | I | ||||||
| IFIH1 | 1426276_at | I | D | |||||||
| IFNAR2 | 1451462_a_at | I | ||||||||
| IGFBP4 | 1421992_a_at, 1423756_s_at, 1423757_x_at, 1437405_a_at, 1437406_x_at | D | D | |||||||
| IGFBP5 | 1422313_a_at, 1452114_s_at | D | D | D | D | |||||
| IGFBP7 | 1423584_at, 1423585_at | D | I | I | ||||||
| IKBKG | 1454690_at | I | ||||||||
| IKIP | 1429065_at, 1429219_at | I | I | |||||||
| IL15 | 1418219_at | I | ||||||||
| IL15RA | 1448681_at | I | ||||||||
| IL18 | 1417932_at | I | I | |||||||
| IL1RL1 | 1422317_a_at | I | ||||||||
| IL6 | 1450297_at | I | I | I | ||||||
| INHBA | 1422053_at | I | I | I | ||||||
| INPP1 | 1418045_at, 1442073_at | I | ||||||||
| IRF8 | 1416714_at, 1448452_at | I | I | |||||||
| IRS1 | 1423104_at | D | I | I | I | |||||
| ITGA5 | 1423267_s_at | D | ||||||||
| ITGB5 | 1417533_a_at, 1417534_at, 1456195_x_at | D | D | |||||||
| ITM2B | 1417999_at, 1418000_a_at | I | I | |||||||
| ITPR3 | 1417297_at | I | I | |||||||
| JMJD6 | 1420056_s_at, 1454109_a_at | D | D | |||||||
| JUN | 1417409_at, 1448694_at | D | D | D | ||||||
| KAT2B | 1434037_s_at, 1450821_at | I | I | |||||||
| KAT5 | 1433980_at, 1433981_s_at | D | ||||||||
| KIF1B | 1455182_at | I | ||||||||
| KITLG | 1415855_at, 1448117_at | I | I | |||||||
| KLF10 | 1416029_at | I | I | |||||||
| KLF4 | 1417394_at, 1417395_at | I | I | |||||||
| KLF6 | 1418280_at, 1427742_a_at, 1447448_s_at | D | D | D | I | |||||
| LAMP2 | 1416344_at | I | I | |||||||
| LCN2 | 1427747_a_at | I | I | |||||||
| LDLR | 1421821_at | D | I | I | ||||||
| LGALS3 | 1426808_at | I | ||||||||
| LGALS3BP | 1448380_at | I | I | |||||||
| LGALS8 | 1422662_at | I | ||||||||
| LIF | 1421207_at | I | ||||||||
| LIMS1 | 1418232_s_at | D | ||||||||
| LMNA | 1421654_a_at, 1425472_a_at, 1457670_s_at | D | D | D | I | I | I | |||
| LPAR1 | 1426110_a_at, 1448606_at | D | D | I | ||||||
| LRIG1 | 1434210_s_at, 1449893_a_at | I | I | |||||||
| LTBR | 1416435_at | D | ||||||||
| MAOA | 1428667_at | I | I | I | ||||||
| MAP2K3 | 1451714_a_at | I | ||||||||
| MAP3K12 | 1438908_at | I | ||||||||
| MAP3K4 | 1459800_s_at | I | I | |||||||
| MAP3K7 | 1419988_at | I | ||||||||
| MAPK3 | 1427060_at | D | ||||||||
| MAPK8 | 1420932_at | D | ||||||||
| MAPKAP1 | 1417284_at | I | I | |||||||
| MAX | 1423501_at | D | D | D | ||||||
| MCF2L | 1434140_at | D | ||||||||
| MCL1 | 1416880_at | D | ||||||||
| MCM2 | 1448777_at, 1423605_a_at, 1427718_a_at | D | D | D | ||||||
| MDM2 | 1427718_a_at | I | I | I | I | I | I | |||
| MED1 | 1450402_at | I | ||||||||
| MEF2A | 1427186_a_at, 1452347_at | I | ||||||||
| MET | 1422990_at, 1434447_at | I | I | |||||||
| MFGE8 | 1420911_a_at | I | ||||||||
| MGP | 1448416_at | D | D | I | I | |||||
| MGST1 | 1415897_a_at | I | I | I | ||||||
| MMP2 | 1416136_at | I | ||||||||
| MMP3 | 1418945_at | I | I | |||||||
| MPG | 1417571_at, 1417572_at | I | I | |||||||
| MT1E | 1428942_at | D | I | I | I | |||||
| MT1F | 1422557_s_at | D | I | |||||||
| MTMR6 | 1425485_at | I | ||||||||
| MTPN | 1437457_a_at | I | ||||||||
| MX1 | 1451905_a_at | D | I | |||||||
| MYC | 1424942_a_at | D | ||||||||
| MYO6 | 1433942_at | I | ||||||||
| NAMPT | 1417190_at | D | D | |||||||
| NCAM1 | 1426864_a_at | I | ||||||||
| NCAPG2 | 1417926_at | I | ||||||||
| NDRG1 | 1420760_s_at, 1423413_at, 1450976_at, 1456174_x_at | D | D | D | ||||||
| NDST1 | 1422044_at, 1460436_at | D | D | |||||||
| NDUFAF4 | 1427997_at | I | ||||||||
| NDUFV2 | 1428179_at, 1452692_a_at | I | ||||||||
| NEDD9 | 1422818_at | I | ||||||||
| NEK2 | 1417299_at, 1437580_s_at | I | I | |||||||
| NEK6 | 1423596_at, 1425850_a_at | I | I | |||||||
| NFAT5 | 1438999_a_at, 1439805_at | D | I | I | I | |||||
| NFIL3 | 1418932_at | D | ||||||||
| NFKB1 | 1427705_a_at | I | ||||||||
| NFKB2 | 1425902_a_at | I | ||||||||
| NFKBIA | 1420088_at, 1438157_s_at, 1448306_at, 1449731_s_at | I | I | I | ||||||
| NFKBIZ | 1417483_at, 1448728_a_at, 1457404_at | I | I | I | ||||||
| NGF | 1419675_at | D | ||||||||
| NME1 | 1424110_a_at | D | ||||||||
| NOD1 | 1454733_at | I | I | |||||||
| NOTCH2 | 1455556_at | D | ||||||||
| NP | 1416530_a_at, 1453299_a_at | I | I | |||||||
| NQO1 | 1423627_at | I | I | |||||||
| NQO2 | 1449983_a_at, 1455590_at | I | I | |||||||
| NR2F1 | 1418157_at | D | ||||||||
| NR3C1 | 1421867_at, 1457635_s_at, 1460303_at | I | I | |||||||
| NR4A1 | 1416505_at | D | ||||||||
| NRF1 | 1434627_at | D | ||||||||
| NRP1 | 1418084_at | I | ||||||||
| NT5C3 | 1451050_at | I | I | |||||||
| NTRK3 | 1433825_at | D | D | D | ||||||
| NUAK2 | 1429049_at | I | ||||||||
| NUPR1 | 1419665_a_at | I | ||||||||
| OAS1 | 1424775_at | I | ||||||||
| OAS1B | 1425119_at | D | ||||||||
| OAS3 | 1425374_at | D | I | I | ||||||
| ODC1 | 1437711_x_at | D | ||||||||
| OSGIN1 | 1424022_at | D | ||||||||
| P2RX4 | 1425525_a_at, 1452527_a_at | I | I | |||||||
| P2RX7 | 1439787_at | I | ||||||||
| PA2G4 | 1420142_s_at, 1423060_at, 1435372_a_at | D | D | D | ||||||
| PAFAH1B1 | 1460199_a_at | D | D | |||||||
| PAK1 | 1420980_at, 1450070_s_at | D | I | |||||||
| PAK3 | 1435486_at, 1437318_at | D | I | |||||||
| PALLD | 1427228_at, 1433768_at | D | I | |||||||
| PARK7 | 1416526_a_at, 1456194_a_at | D | D | |||||||
| PARVA | 1431375_s_at | I | I | |||||||
| PARVB | 1438672_at | I | ||||||||
| PAWR | 1426910_at | D | I | |||||||
| PCNA | 1417947_at | D | D | |||||||
| PDCD2 | 1423534_at | D | ||||||||
| PDGFRA | 1421917_at | D | D | |||||||
| PDGFRB | 1417148_at, 1436970_a_at | D | D | D | D | |||||
| PEA15 | 1416407_at | I | ||||||||
| PHLDA1 | 1418835_at | I | I | I | ||||||
| PIK3CA | 1460326_at | I | ||||||||
| PIK3R2 | 1418463_at | I | ||||||||
| PITPNA | 1423282_at, 1423283_at | I | I | |||||||
| PKN2 | 1437295_at, 1437296_at | I | ||||||||
| PLAC8 | 1451335_at | I | I | |||||||
| PLAT | 1415806_at | I | I | |||||||
| PLAUR | 1452521_a_at | I | ||||||||
| PLD1 | 1437113_s_at | I | I | |||||||
| PLD2 | 1417237_at | I | ||||||||
| PLEKHF1 | 1424671_at | I | I | |||||||
| PLK1 | 1448191_at | D | I | I | I | |||||
| PLK3 | 1434496_at | I | I | |||||||
| PLSCR1 | 1429527_a_at, 1453181_x_at | I | I | |||||||
| PLSCR3 | 1449020_at | I | ||||||||
| PMEPA1 | 1422706_at, 1452295_at | D | D | D | ||||||
| PML | 1448757_at, 1456103_at | D | I | |||||||
| PNKP | 1416378_at | I | I | |||||||
| PNPT1 | 1452676_a_at | D | ||||||||
| POLK | 1449483_at | I | I | |||||||
| PPID | 1417057_a_at | D | D | |||||||
| PPM1A | 1429501_s_at, 1451943_a_at | D | ||||||||
| PPM1F | 1454934_at | I | ||||||||
| PPP1R13L | 1459592_a_at | D | ||||||||
| PPP1R15A | 1448325_at | D | D | |||||||
| PPP2R2A | 1437730_at, 1453260_a_at | D | D | |||||||
| PRDX5 | 1416381_a_at | I | ||||||||
| PRKAR2B | 1438664_at, 1456475_s_at | I | ||||||||
| PRKCA | 1450945_at | I | ||||||||
| PRKD1 | 1447623_s_at | I | ||||||||
| PRMT2 | 1416844_at | I | ||||||||
| PRPF19 | 1460633_at | D | ||||||||
| PRR13 | 1423686_a_at | I | I | I | ||||||
| PSENEN | 1415679_at | D | ||||||||
| PSIP1 | 1417166_at, 1460403_at | I | I | I | ||||||
| PSMG2 | 1425373_a_at, 1448212_at | D | ||||||||
| PTGR1 | 1417777_at | I | I | I | ||||||
| PTGS1 | 1436448_a_at | I | ||||||||
| PTGS2 | 1417262_at, 1417263_at | I | I | I | ||||||
| PTMA | 1423455_at | D | D | |||||||
| PTPN1 | 1438670_at | D | ||||||||
| PTPRA | 1425340_a_at | I | I | |||||||
| PTPRE | 1418540_a_at | I | ||||||||
| PTPRG | 1434360_s_at | D | D | |||||||
| PTRH2 | 1451845_a_at | D | D | D | ||||||
| PTTG1 | 1419620_at, 1424105_a_at, 1438390_s_at | I | I | I | ||||||
| PXN | 1424027_at, 1456135_s_at | I | I | |||||||
| QARS | 1423712_a_at, 1456726_x_at | I | I | |||||||
| QKI | 1417073_a_at, 1425597_a_at, 1429318_a_at, 1451179_a_at | D | D | D | D | |||||
| RABGGTB | 1419553_a_at | I | ||||||||
| RAD18 | 1451928_a_at | I | ||||||||
| RAD21 | 1416162_at | D | ||||||||
| RAD54L | 1450862_at | I | I | I | ||||||
| RALB | 1417744_a_at | I | ||||||||
| RARG | 1419415_a_at, 1419416_a_at | D | ||||||||
| RASA1 | 1426476_at, 1426477_at | I | ||||||||
| RASSF1 | 1441737_s_at, 1448855_at | I | ||||||||
| RASSF5 | 1422637_at | I | ||||||||
| RB1 | 1417850_at | I | ||||||||
| RBBP4 | 1434892_x_at, 1454791_a_at, 1454875_a_at | D | D | D | ||||||
| RBBP6 | 1425114_at | D | ||||||||
| RBL1 | 1424156_at, 1425166_at | D | D | |||||||
| RBP1 | 1448754_at | I | I | I | ||||||
| RCAN2 | 1421425_a_at | I | ||||||||
| RECK | 1450784_at | I | ||||||||
| RFC1 | 1418342_at, 1449050_at, 1451920_a_at | I | I | I | ||||||
| RFK | 1415737_at, 1416230_at | D | D | |||||||
| RFWD2 | 1426913_at | I | ||||||||
| RGS3 | 1425296_a_at, 1425701_a_at | I | I | I | ||||||
| RIPK1 | 1419508_at, 1449485_at | I | I | |||||||
| RIPK2 | 1450173_at | I | ||||||||
| RNF34 | 1415791_at | I | ||||||||
| ROCK1 | 1423444_at, 1423445_at | I | I | |||||||
| RPS3 | 1435151_a_at | I | ||||||||
| RPS3A | 1422475_a_at | I | I | |||||||
| RPS6KB1 | 1454956_at | I | ||||||||
| RRAS | 1418448_at | I | ||||||||
| RRAS2 | 1417398_at | I | ||||||||
| RRM2B | 1437476_at | I | ||||||||
| RTN4 | 1421116_a_at, 1452649_at | D | I | |||||||
| S100A1 | 1417421_at, 1419814_s_at | I | I | |||||||
| S100A10 | 1416762_at, 1456642_x_at | I | I | I | ||||||
| S100A4 | 1424542_at | D | D | I | I | I | ||||
| S100A6 | 1421375_a_at | I | I | |||||||
| S1PR1 | 1423571_at | D | I | I | ||||||
| S1PR2 | 1428176_at | D | ||||||||
| S1PR3 | 1438658_a_at | I | ||||||||
| SAT1 | 1420502_at | I | ||||||||
| SCARB1 | 1416050_a_at, 1437378_x_at, 1455820_x_at | I | I | |||||||
| SDC1 | 1415943_at, 1415944_at, 1437279_x_at | D | D | I | ||||||
| SDC4 | 1448793_a_at | I | ||||||||
| SEMA3A | 1449865_at | I | ||||||||
| SENP1 | 1424330_at | D | ||||||||
| SERBP1 | 1437280_s_at | I | ||||||||
| SERPINE1 | 1419149_at | D | D | I | I | I | ||||
| SERPINE2 | 1416666_at | I | ||||||||
| SERPINF1 | 1416168_at, 1453724_a_at | D | ||||||||
| SFRP1 | 1448395_at | I | ||||||||
| SFRP2 | 1448201_at | D | ||||||||
| SFRS5 | 1423130_a_at | I | ||||||||
| SGK1 | 1416041_at | I | I | I | ||||||
| SGMS2 | 1428663_at, 1429029_at | I | ||||||||
| SGPL1 | 1415892_at | D | ||||||||
| SH3BP5 | 1421922_at, 1421923_at | I | I | |||||||
| SH3GLB1 | 1418011_a_at, 1418012_at | D | I | |||||||
| SH3KBP1 | 1431592_a_at, 1460337_at | I | I | |||||||
| SHISA5 | 1423986_a_at, 1437503_a_at | I | I | |||||||
| SHPRH | 1452261_at | I | ||||||||
| SIRT7 | 1424238_at | I | ||||||||
| SKIL | 1452214_at | I | ||||||||
| SLC25A24 | 1427483_at, 1452717_at | I | I | |||||||
| SLC2A1 | 1426599_a_at, 1434773_a_at | D | I | |||||||
| SLC7A11 | 1420413_at | I | ||||||||
| SLK | 1425977_a_at, 1449336_a_at | I | ||||||||
| SMN1 | 1426596_a_at | D | D | |||||||
| SMNDC1 | 1429043_at | D | ||||||||
| SNRPE | 1451294_s_at | D | D | |||||||
| SOCS3 | 1416576_at, 1455899_x_at, 1456212_x_at | D | I | I | I | |||||
| SOD2 | 1417193_at, 1448610_a_at | I | ||||||||
| SOD3 | 1417633_at | I | I | |||||||
| SORBS2 | 1437197_at | I | I | I | ||||||
| SOX4 | 1419155_a_at, 1419156_at, 1419157_at, 1433575_at, 1449370_at | D | D | D | D | D | ||||
| SP1 | 1418180_at, 1454852_at | D | D | |||||||
| SPP1 | 1449254_at | D | D | I | I | I | ||||
| SRGN | 1417426_at | I | I | |||||||
| STAT1 | 1420915_at, 1450033_a_at, 1450034_at | D | D | D | ||||||
| STAT5A | 1421469_a_at, 1450259_a_at | I | I | |||||||
| STAT6 | 1426353_at | I | I | |||||||
| STK24 | 1426248_at | D | ||||||||
| STMN1 | 1415849_s_at, 1448113_at | D | D | |||||||
| STX8 | 1418089_at | I | I | |||||||
| SULF1 | 1436319_at, 1438200_at | I | ||||||||
| TACC3 | 1417450_a_at, 1436872_at, 1455834_x_at | I | I | |||||||
| TADA3L | 1417467_a_at | I | ||||||||
| TAX1BP1 | 1420174_s_at, 1448399_at | I | I | |||||||
| TCF12 | 1427670_a_at | D | ||||||||
| TCF4 | 1416724_x_at | I | ||||||||
| TCF7 | 1433471_at | I | ||||||||
| TENC1 | 1452264_at | D | I | |||||||
| TERF1 | 1418380_at | I | ||||||||
| TFAP2A | 1421996_at, 1426048_s_at | D | D | |||||||
| TGFB1 | 1420653_at | I | I | |||||||
| TGFB1I1 | 1418136_at | I | ||||||||
| TGFB2 | 1450922_a_at | I | I | |||||||
| TGFBR2 | 1425444_a_at, 1426397_at | I | I | |||||||
| TGFBR3 | 1433795_at | D | I | |||||||
| THBS1 | 1421811_at, 1450377_at, 1460302_at | D | I | I | I | |||||
| THBS2 | 1422571_at, 1447862_x_at, 1450663_at | D | D | D | I | |||||
| TIAL1 | 1421148_a_at | D | ||||||||
| TIMP1 | 1460227_at | I | I | |||||||
| TIMP2 | 1420924_at, 1433662_s_at, 1450040_at, 1454677_at, 1460287_at | I | I | I | ||||||
| TIMP3 | 1419088_at, 1419089_at, 1449334_at, 1449335_at | D | I | I | ||||||
| TLR1 | 1449049_at | I | ||||||||
| TLR3 | 1422781_at, 1422782_s_at | I | D | I | I | |||||
| TLR4 | 1418163_at | I | ||||||||
| TMEM173 | 1427911_at, 1447621_s_at | I | I | I | ||||||
| TMSB10 | 1417219_s_at, 1436902_x_at, 1437185_s_at | I | I | |||||||
| TMSB4X | 1415906_at | I | I | I | ||||||
| TNC | 1416342_at, 1456344_at | D | I | |||||||
| TNFAIP3 | 1433699_at | I | I | |||||||
| TNFAIP8 | 1416950_at | I | I | |||||||
| TNFRSF12A | 1418571_at, 1418572_x_at | I | I | I | ||||||
| TNFRSF19 | 1425212_a_at | I | ||||||||
| TNFRSF1A | 1417291_at | D | ||||||||
| TNKS2 | 1447522_s_at | I | I | |||||||
| TOP1 | 1423474_at | I | ||||||||
| TOP2A | 1454694_a_at | I | I | |||||||
| TOPBP1 | 1452241_at | I | I | |||||||
| TOPORS | 1417754_at | I | ||||||||
| TP53 | 1426538_a_at, 1427739_a_at | D | ||||||||
| TP53BP2 | 1433937_at, 1433938_at | D | D | |||||||
| TP53INP1 | 1416926_at, 1416927_at | I | I | I | I | I | I | |||
| TPD52L1 | 1418412_at | I | ||||||||
| TPM1 | 1423049_a_at, 1423721_at | I | I | |||||||
| TPP1 | 1434768_at | I | I | |||||||
| TRAF3IP2 | 1448508_at | I | ||||||||
| TRAF7 | 1424320_a_at | I | I | |||||||
| TRIAP1 | 1460702_at | D | ||||||||
| TRIB2 | 1426640_s_at | D | D | |||||||
| TRIB3 | 1426065_a_at, 1456225_x_at | D | D | D | ||||||
| TRIM27 | 1438376_s_at, 1456375_x_at | D | ||||||||
| TSC2 | 1452105_a_at | I | ||||||||
| TSLP | 1450004_at | I | I | |||||||
| TSPO | 1416695_at, 1438948_x_at, 1456251_x_at | I | I | |||||||
| TTK | 1449171_at | I | I | I | ||||||
| TXN | 1416119_at | I | ||||||||
| TXNDC17 | 1423034_at, 1423035_s_at, 1439184_s_at | I | I | |||||||
| TXNIP | 1415996_at, 1415997_at | I | D | D | ||||||
| UBA7 | 1426971_at | I | I | |||||||
| UBE2C | 1452954_at | I | I | I | ||||||
| UBR4 | 1454668_at | D | ||||||||
| UNG | 1425753_a_at | D | D | D | D | D | ||||
| UTP11L | 1429485_a_at | I | ||||||||
| UXT | 1418986_a_at | D | ||||||||
| VCAM1 | 1415989_at, 1436003_at, 1448162_at, 1451314_a_at | I | I | |||||||
| VCAN | 1427256_at | D | ||||||||
| VCL | 1416156_at, 1416157_at | I | I | |||||||
| VDR | 1418175_at, 1418176_at | I | ||||||||
| VHL | 1434708_at | D | D | |||||||
| WEE1 | 1416773_at | D | ||||||||
| WFS1 | 1448411_at | D | ||||||||
| WISP1 | 1448593_at, 1448594_at | I | I | |||||||
| WRN | 1425982_a_at | I | ||||||||
| WTAP | 1454805_at | D | D | |||||||
| WWOX | 1416334_at | D | D | |||||||
| XAF1 | 1443698_at | I | ||||||||
| XBP1 | 1420886_a_at, 1437223_s_at | D | ||||||||
| XDH | 1451006_at | I | ||||||||
| XPA | 1460725_at | I | ||||||||
| XRCC2 | 1455335_at | D | ||||||||
| XRCC4 | 1424601_at | D | D | |||||||
| XRCC6 | 1417437_at | D | D | |||||||
| YARS | 1460638_at | D | ||||||||
| YWHAE | 1435702_s_at, 1438839_a_at | D | ||||||||
| YY1 | 1435824_at, 1457834_at | D | I | I | ||||||
| ZFP36 | 1452519_a_at | D | I | |||||||
| ZFP36L2 | 1437626_at | D | D | I | ||||||
| ZMAT3 | 1449353_at | I | I | I | ||||||
| ZNF148 | 1418381_at, 1449068_at, 1449069_at | I | I | |||||||
| ZNF622 | 1438000_x_at | D | D | |||||||
| ZYX | 1417240_at | I | I | I | ||||||
Figure 3.
Microarray findings are summarized by Venn diagrams which show the distribution of differentially expressed probeset IDs in each treatment group [irradiation (Gy), cisplatin (CDDP) or combination of both (Gy and CDDP)] when compared to nontreated control cells at 3 h, 24 h, and 72 h after treatment.
Table 2.
Differential expression of apoptosis-related genes which have direct upstream or downstream relationship with p53 in each treatment group [irradiation (Gy), cisplatin (CDDP), or combination of both (Gy + CDDP)] when compared to nontreated control cells at 3 h, 24 h, and 72 h after treatment.
| 3 hours | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Symbol | Gy | CDDP | Gy+CDDP | Symbol | Gy | CDDP | Gy+CDDP |
| CCNG1 | I | I | KLF6 | D | D | D | |
| CDKN1A | I | I | CCND1 | D | |||
| MDM2 | I | I | CTCF | D | |||
| TP53INP1 | I | I | IRS1 | D | |||
| BTG2 | I | ATF3 | D | ||||
| DDX5 | I | AURKA | D | ||||
| GDF15 | I | BID | D | ||||
| IL6 | I | CCNB1 | D | ||||
| C11ORF82 | I | CEBPB | D | ||||
| CASP3 | I | DDR1 | D | ||||
| CASP6 | I | ETS1 | D | ||||
| CRYAB | I | FHL2 | D | ||||
| HSP90AA1 | I | HBEGF | D | ||||
| MED1 | I | HIPK1 | D | ||||
| ZMAT3 | I | HSPB1 | D | ||||
| MAPK3 | D | ||||||
| MCL1 | D | ||||||
| MYC | D | ||||||
| NR4A1 | D | ||||||
| OSGIN1 | D | ||||||
| PLK1 | D | ||||||
| PMEPA1 | D | ||||||
| PPP1R13L | D | ||||||
| THBS1 | D | ||||||
| THBS2 | D | ||||||
| YY1 | D | ||||||
|
| |||||||
| 24 hours | |||||||
|
| |||||||
| Symbol | Gy | CDDP | Gy+CDDP | Symbol | Gy | CDDP | Gy+CDDP |
|
| |||||||
| CCNG1 | I | I | I | APEX1 | D | D | |
| CDKN1A | I | I | I | BHLHE40 | D | D | |
| EIF2AK2 | I | I | I | BRE | D | D | |
| RFC1 | I | I | I | PMEPA1 | D | D | |
| BUB1 | I | I | S100A4 | D | D | ||
| CCAR1 | I | I | SERPINE1 | D | D | ||
| CKAP2 | I | I | SPP1 | D | D | ||
| HSP90AA1 | I | I | THBS2 | D | D | ||
| MDM2 | I | I | WWOX | D | D | ||
| TOP2A | I | I | SLC2A1 | D | |||
| TP53INP1 | I | I | BTG1 | D | |||
| ZNF148 | I | I | CAV1 | D | |||
| KLF6 | I | CEBPB | D | ||||
| BLM | I | TIMP3 | D | ||||
| BRCA1 | I | ATP1A1 | D | ||||
| HMMR | I | CDK4 | D | ||||
| IFI16 | I | CDKN2A | D | ||||
| RAD54L | I | CDKN2C | D | ||||
| RB1 | I | GSTM5 | D | ||||
| TOP1 | I | ID2 | D | ||||
| TOPBP1 | I | TFAP2A | D | ||||
| TTK | I | ||||||
| BTG2 | I | ||||||
| C11ORF82 | I | ||||||
| FUBP1 | I | ||||||
| NFKB2 | I | ||||||
| NUPR1 | I | ||||||
| TOPORS | I | ||||||
|
| |||||||
| 72 hours | |||||||
|
| |||||||
| Symbol | Gy | CDDP | Gy+CDDP | Symbol | Gy | CDDP | Gy+CDDP |
|
| |||||||
| ANXA1 | I | I | I | LGALS3 | I | ||
| AURKA | I | I | I | LIF | I | ||
| BUB1B | I | I | I | MAP2K3 | I | ||
| CAV1 | I | I | I | MMP2 | I | ||
| CCNB1 | I | I | I | MYO6 | I | ||
| CCND1 | I | I | I | NFKB1 | I | ||
| CCNG1 | I | I | I | PLAUR | I | ||
| CDC20 | I | I | I | PRKCA | I | ||
| CDKN1A | I | I | I | PTGS1 | I | ||
| CLU | I | I | I | SAT1 | I | ||
| DDIT4 | I | I | I | SERPINE2 | I | ||
| EGR1 | I | I | I | SLC2A1 | I | ||
| FOSL1 | I | I | I | SOD2 | I | ||
| GLIPR1 | I | I | I | TADA3L | I | ||
| HSPB1 | I | I | I | THBS2 | I | ||
| IER3 | I | I | I | TOPBP1 | I | ||
| INHBA | I | I | I | TSC2 | I | ||
| IRS1 | I | I | I | VDR | I | ||
| NFKBIA | I | I | I | FEN1 | D | D | D |
| PHLDA1 | I | I | I | GADD45A | D | D | D |
| PLK1 | I | I | I | JUN | D | D | D |
| PTGS2 | I | I | I | MCM2 | D | D | D |
| PTTG1 | I | I | I | NDRG1 | D | D | D |
| S100A4 | I | I | I | STAT1 | D | D | D |
| SERPINE1 | I | I | I | ATF3 | D | D | |
| SGK1 | I | I | I | PPP2R2A | D | D | |
| SPP1 | I | I | I | TP53BP2 | D | D | |
| THBS1 | I | I | I | CDK4 | D | D | |
| TMSB4X | I | I | I | FUBP1 | D | D | |
| UBE2C | I | I | I | GSK3B | D | D | |
| ZYX | I | I | I | HMGB1L1 | D | D | |
| BCL3 | I | I | HSPA5 | D | D | ||
| MMP3 | I | I | PARK7 | D | D | ||
| S100A6 | I | I | PCNA | D | D | ||
| ABCB1B | I | I | PPP1R15A | D | D | ||
| AKAP12 | I | I | SMN1 | D | D | ||
| ATM | I | I | SP1 | D | D | ||
| BID | I | I | STMN1 | D | D | ||
| BIRC5 | I | I | XRCC6 | D | D | ||
| BUB1 | I | I | DAXX | D | I | ||
| CCNA2 | I | I | MX1 | D | I | ||
| CCND3 | I | I | PML | D | I | ||
| CDC25C | I | I | E2F1 | D | |||
| CKAP2 | I | I | IFI16 | D | |||
| CRYAB | I | I | BRE | D | |||
| CTSD | I | I | CDC6 | D | |||
| DDR1 | I | I | CHEK1 | D | |||
| DHCR24 | I | I | COPS5 | D | |||
| EZR | I | I | CTCF | D | |||
| FHL2 | I | I | DDIT3 | D | |||
| FOS | I | I | DUT | D | |||
| FOXM1 | I | I | ELAVL1 | D | |||
| HMMR | I | I | HOXA7 | D | |||
| IL6 | I | I | HUWE1 | D | |||
| KAT2B | I | I | KAT5 | D | |||
| KLF4 | I | I | MAPK8 | D | |||
| MDM2 | I | I | NME1 | D | |||
| MET | I | I | RBBP6 | D | |||
| NEK2 | I | I | TFAP2A | D | |||
| NQO1 | I | I | TP53 | D | |||
| NQO2 | I | I | VCAN | D | |||
| NR3C1 | I | I | |||||
| PLK3 | I | I | |||||
| PTPRA | I | I | |||||
| RAD54L | I | I | |||||
| S100A1 | I | I | |||||
| SHISA5 | I | I | |||||
| TACC3 | I | I | |||||
| TGFB2 | I | I | |||||
| TIMP3 | I | I | |||||
| TP53INP1 | I | I | |||||
| TTK | I | I | |||||
| YY1 | I | I | |||||
| ZMAT3 | I | I | |||||
| TXN | I | ||||||
| GDF15 | I | ||||||
| GSTM1 | I | ||||||
| RFWD2 | I | ||||||
| RRM2B | I | ||||||
| WRN | I | ||||||
| AFP | I | ||||||
| AHR | I | ||||||
| AP2A2 | I | ||||||
| BHLHE40 | I | ||||||
| C11ORF82 | I | ||||||
| CASP6 | I | ||||||
| CAT | I | ||||||
| CDK8 | I | ||||||
| CENPF | I | ||||||
| CFLAR | I | ||||||
| CSF2 | I | ||||||
| CX3CL1 | I | ||||||
| EPHA2 | I | ||||||
| ERCC3 | I | ||||||
| ERCC5 | I | ||||||
| ETS1 | I | ||||||
| FAS | I | ||||||
| FASN | I | ||||||
| GPI | I | ||||||
| HK2 | I | ||||||
| HSP90AB1 | I | ||||||
| ID1 | I | ||||||
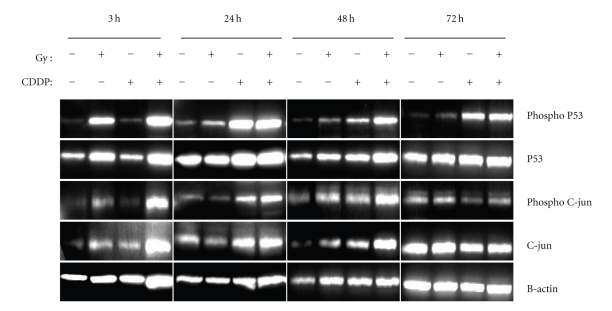
3.4. Differential Temporal Activation of p53 Occurred with CDDP and Radiation Treatment
It was observed that Posttreatment activation of p53 occurred earlier in radiation than in CDDP (Figure 4). In response to DNA damage, activation of the p53 pathway normally occurs with the phosphorylation of ser-15 in p53. The present study showed radiation-induced phosphorylation of p53 occurred at 3 hrs after treatment, compared to CDDP-induced activation which was observed only at 24 hrs or later (Figure 4). These timings corresponded with those observed for the expression of apoptotic-related genes after radiation and CDDP treatment (Figure 3). For example, MDM2 and TP53INP1 were expressed at 3 hrs after radiation. They were however, expressed only at 24 hrs after CDDP (Table 2).
Figure 4.
Western blot analysis showing p53 and c-jun protein expression and phosphorylation at various time points (3 h, 24 h, 48 h, and 72 h) after 5 Gy of gamma radiation and 0.5 μg/ml pf cisplatin (CDDP). The data are representative of 3 separate experiments.
4. Discussion
Combined chemoradiation is increasingly being used to treat advanced head and neck caners. As radiation and CDDP are both ototoxic, it is of concern that significant sensorineural hearing loss will result. Indeed, patients with nasopharyngeal carcinoma who had received radiotherapy and concurrent/adjuvant chemotherapy using CDDP were found to experience greater sensorineural hearing loss compared with patients treated with radiotherapy alone, especially to high-frequency sounds in the speech range [1]. It is of interest to note that different etiologies of sensorineural hearing loss, such as noise, ototoxic drugs, and aging, result in similar patterns of audiometric changes and cochlear cellular degeneration [8]. The cellular and molecular mechanisms involved in sensorineural hearing loss from diverse causes appear to lead to a final common pathway which results in apoptosis of cochlear hair cells [6, 9].
In radiation-induced ototoxicity, cochlear cell apoptosis and ROS generation were observed after irradiation, and p53 was thought to play a key role [7]. This phenomenon was dose dependant and occurred predominantly at 72 h after irradiation. Microarray analysis supported these findings, as associated dose-dependant apoptotic gene regulation changes were observed.
The ototoxic manifestations of CDDP are primarily due to its effects on the cochlear hair cells although the spiral ganglion cells and the stria vascularis are also affected to some extent. According to Rybak et al. [10], CDDP ototoxicity appears to be triggered by ROSs that initiate a cascade of molecular events that lead to apoptosis of outer hair cells, resulting in hearing loss. Ototoxic effects on the stria vascularis are transient, resulting in temporary reduction of endocochlear potential associated with stria edema. The endocochlear potential recovers but residual shrinkage of the strial persists. The spiral ganglia are thought to be least affected.
Although the cellular and molecular processes of ototoxicity have been described for radiation and CDDP when used alone, those involved in combined therapy have not been studied previously. The present study demonstrated that combined therapy led to decreased viability of cochlear cells, with an increase in the subG1 population. These findings support the belief that as in other etiologies of sensorineural loss, apoptosis of cochlear hair cells is important in CDDP-radiation.
It is well established that p53 plays a key role in the cellular response to nuclear DNA damage [11]. It regulates cell cycle arrest and dictates cell fate like senescence, apoptosis, and DNA repair. It is believed that the nature of DNA damage enables p53 to selectively discriminate between promotors in the induction of target genes, thereby regulating their expression and subsequent cellular outcome [12].
In a study on HEI-OC1 cells derived from the cochlea, CDDP caused an increase in p53 at 3 hrs prior to the activation of Bax, cytochrome-c, and caspase 8 and 9 [13]. In the case of radiation-induced ototoxicity, the role of p53 in triggering apoptotic cell death in cochlear hair cells has also been studied [7]. Based on microarray analysis, the p53 gene was found to be up-regulated after irradiation and p53 expression was confirmed by Western blotting. Although p53 plays a role in both CDDP and radiation-induced ototoxicity, the present study showed that p53 was activated at different time points after treatment. Posttreatment phosphorylation of p53 occurred after 24 hrs for CDDP, whereas it occurred as early as 3 hrs for radiation. These timings corresponded to the times MDM2 and TP53INP1 were expressed after treatment with CDDP and radiation respectively. Therefore, although both CDDP and radiation-induced cochlear cell apoptosis appear to involve activation of p53, the upstream processes involved may well be different.
In the present study, combined CDDP-radiation treatment triggered more apoptotic-related gene expressions than those that could be accounted for by a summation of gene expressions resulting from individual treatments. This could explain the synergistic ototoxic effects of combined CDDP-radiation treatment, an observation seen clinically [1]. Interestingly, among the genes which were expressed in combined treatment but not when these entities were used alone was FAS, a key element involved in the extrinsic apoptotic pathway. Although the extrinsic apoptotic pathway has generally been regarded to play only minor role in ototoxicity resulting from the use of CDDP or radiation alone, it may well be important in situations when they are used in combination [14, 15].
The OC-k3 cell line expressed the neuroepithelial precursor cell marker nestin and the inner ear cell marker OCP2, specific auditory sensory cell markers myosin VIIa and the acetylcholine receptor alpha-9 and the supporting cell marker connexin 26. It had been regarded as a good model to study the mechanisms of cell fate in the Organ of Corti of the cochlea [4]. Therefore, the finding that combined treatment actually led to enhanced apoptotic gene expressions including FAS should be further investigated in in vivo animal studies which may have implications in future antiapoptotic treatments against ototoxicity.
5. Conclusion
Like in other etiologies of sensorineural loss, apoptosis of cochlear hair cells appears to play a role in ototoxicity resulting from combined CDDP-radiation therapy. Differential temporal activation of p53 suggests the possibility of different upstream processes leading to its activation after CDDP and radiation treatment. Enhanced apoptotic gene expressions including that of FAS were observed in combined treatment which could possibly explain the synergistic ototoxic effects seen clinically.
Acknowledgments
This study has been supported by a grant from the Department of Clinical Research, Singapore General Hospital. The authors thank Dr F. Kalinec (House Ear Institute, LA, USA) for providing the cell line and Mr Alvin WC Chua (Department of Plastic Reconstructive & Aesthetic Surgery, Singapore General Hospital) for his guidance in cell culture.
References
- 1.Low WK, Toh ST, Wee J, Fook-Chong SMC, Wang DY. Sensorineural hearing loss after radiotherapy and chemoradiotherapy: a single, blinded, randomized study. Journal of Clinical Oncology. 2006;24(12):1904–1909. doi: 10.1200/JCO.2005.05.0096. [DOI] [PubMed] [Google Scholar]
- 2.Rivolta MN, Holley MC. Cell lines in inner ear research. Journal of Neurobiology. 2002;53(2):306–318. doi: 10.1002/neu.10111. [DOI] [PubMed] [Google Scholar]
- 3.Kalinec F, Kalinec G, Boukhvalova M, Kachar B. Establishment and characterization of conditionally immortalized organ of Corti cell lines. Cell Biology International. 1999;23(3):175–184. doi: 10.1006/cbir.1998.0339. [DOI] [PubMed] [Google Scholar]
- 4.Kalinec GM, Webster P, Lim DJ, Kalinec F. A cochlear cell line as an in vitro system for drug ototoxicity screening. Audiology and Neuro-Otology. 2003;8(4):177–189. doi: 10.1159/000071059. [DOI] [PubMed] [Google Scholar]
- 5.Zhang M, Liu W, Ding D, Salvi R. Pifithrin-α supresses p53 and protects cochlear and vestibular hair cells from cisplatin-induced apoptosis. Neuroscience. 2003;120(1):191–205. doi: 10.1016/s0306-4522(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AG, Cunningham LL, Rubel EW. Mechanisms of hair cell death and protection. Current Opinion in Otolaryngology and Head and Neck Surgery. 2005;13(6):343–348. doi: 10.1097/01.moo.0000186799.45377.63. [DOI] [PubMed] [Google Scholar]
- 7.Low W-K, Tan MGK, Sun L, Chua AWC, Goh L-K, Wang D-Y. Dose-dependant radiation-induced apoptosis in a cochlear cell-line. Apoptosis. 2006;11(12):2127–2136. doi: 10.1007/s10495-006-0285-4. [DOI] [PubMed] [Google Scholar]
- 8.Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise-induced hearing loss. Ear and Hearing. 2006;27(1):1–19. doi: 10.1097/01.aud.0000191942.36672.f3. [DOI] [PubMed] [Google Scholar]
- 9.Atar O, Avraham KB. Therapeutics of hearing loss: expectations vs reality. Drug Discovery Today. 2005;10(19):1323–1330. doi: 10.1016/S1359-6446(05)03618-4. [DOI] [PubMed] [Google Scholar]
- 10.Rybak LP, Whitworth CA, Mukherjea D, Ramkumar V. Mechanisms of cisplatin-induced ototoxicity and prevention. Hearing Research. 2007;226(1-2):157–167. doi: 10.1016/j.heares.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Bristow RG, Benchimol S, Hill RP. The p53 gene as a modifier of intrinsic radiosensitivity: implications for radiotherapy. Radiotherapy and Oncology. 1996;40(3):197–223. doi: 10.1016/0167-8140(96)01806-3. [DOI] [PubMed] [Google Scholar]
- 12.Hill R, Bodzak E, Blough MD, Lee PWK. p53 binding to the p21 promoter is dependent on the nature of DNA damage. Cell Cycle. 2008;7(16):2535–2543. doi: 10.4161/cc.7.16.6440. [DOI] [PubMed] [Google Scholar]
- 13.Devarajan P, Savoca M, Castaneda MP, et al. Cisplatin-induced apoptosis in auditory cells: role of death receptor and mitochondrial pathways. Hearing Research. 2002;174(1-2):45–54. doi: 10.1016/s0378-5955(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Ladrech S, Pujol R, Brabet P, Van De Water TR, Puel J-L. Caspase inhibitors, but not c-Jun NH2-terminal kinase inhibitor treatment, prevent cisplatin-induced hearing loss. Cancer Research. 2004;64(24):9217–9224. doi: 10.1158/0008-5472.CAN-04-1581. [DOI] [PubMed] [Google Scholar]
- 15.Verheij M, Bartelink H. Radiation-induced apoptosis. Cell and Tissue Research. 2000;301(1):133–142. doi: 10.1007/s004410000188. [DOI] [PubMed] [Google Scholar]