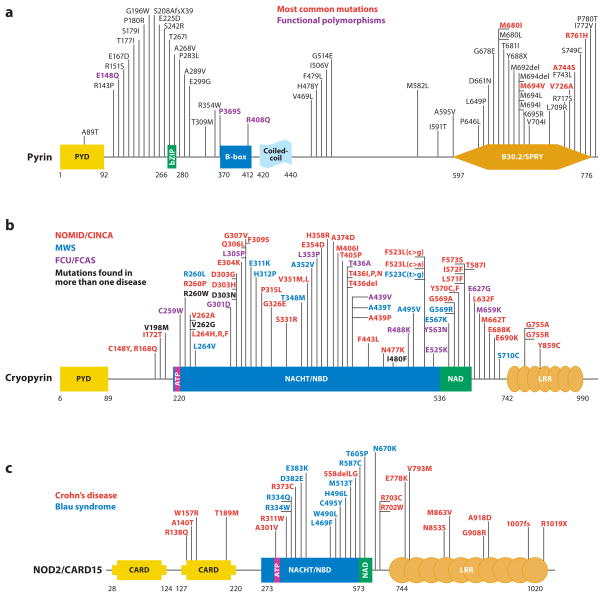
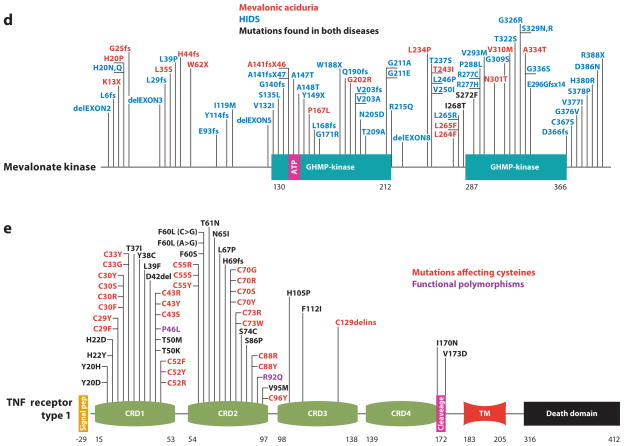
Figure 1.
Schematic representation of mutations in five proteins that cause autoinflammatory diseases. (a) For pyrin, mutations that are most frequently found to cause FMF are presented in red, while those in black are less common disease-causing variants. Residues in purple are found at approximately 1% allele frequency in the general population and may therefore represent functional polymorphisms. (b) Cryopyrin mutations cause a spectrum of disease states that range in severity from severe (NOMID/CINCA, red ), to intermediate (MWS, blue), to mild (FCAS, purple). In some instances, it is difficult to distinguish between NOMID/CINCA and MWS. Residues for which disease presentation overlaps are depicted in black. (c) NOD2/CARD15 mutations can cause Blau syndrome/early-onset sarcoidosis (blue), which cluster within the NACHT/NBD and NAD domains. Other variants that are spread throughout this protein are associated with Crohn’s disease (red ). (d ) Mevalonate kinase mutations can cause the severe metabolic disease mevalonic aciduria (red ), or the less severe autoinflammatory disease HIDS (blue). HIDS mutations are recessively inherited and often include one mild mutation coupled with a severe mutation; thus, the severe mutations can be found in both HIDS and mevalonic aciduria (black). (e) Dominantly inherited missense mutations in TNFR1 that cause TRAPS are now known to affect almost every cysteine residue within the first two cysteine-rich domains of the extracellular region of the protein (red ). These mutations appear to affect the protein folding, whereas at least two mutations, I170N and V173D, can affect ectodomain cleavage that generates the soluble form of the receptor. P46L and R92Q ( purple) are probably functional polymorphisms also present in unaffected individuals. The numbering system for TNFR1 used here begins at residue 30, which is at the N terminus after removal of the 29-residue signal peptide, as per common convention. Abbreviations used: PYD, pyrin domain; NBD, nucleotide-binding domain; NAD, NACHT-associated domain; LRR, leucine-rich repeats; CRD, cysteine-rich domain; TM, transmembrane domain.