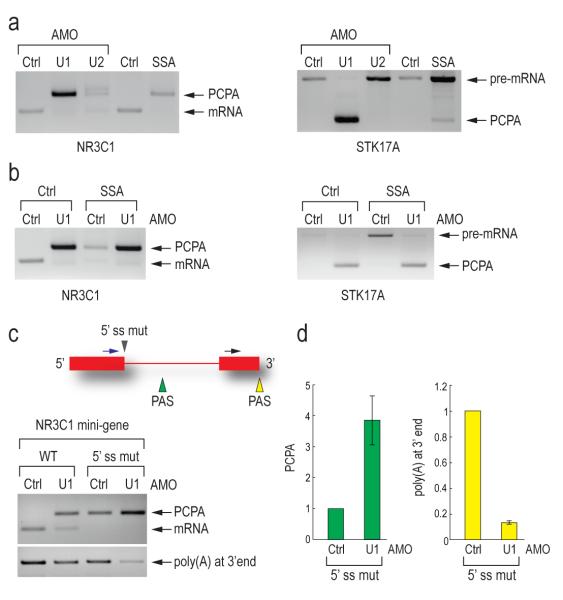
Figure 5. U1 snRNP suppression of premature cleavage and polyadenylation from a nearby cryptic PAS is splicing independent and requires base pairing.
(a) 3′ RACE was carried out as described in Figure 4a on the endogenous NR3C1 and STK17A genes using RNA samples from HeLa cells transfected with control, U1 (7.5 μM, 8 hrs) or U2 AMO (25 μM, 8 hrs), or treated with SSA (100 ng/ml, 8hrs).
(b) 3′ RACE was carried out as described in Figure 4a using RNA samples from HeLa cells transfected with control or U1 AMOs (7.5 μM) with or without SSA (100 ng/ml) for 8 hrs.
(c) HeLa cells were transfected with wild-type and 5′ splice site mutant NR3C1 (mutated from AAGGTAAGA to GTCCATTCA) mini-gene. 3′ RACE was performed as described in Figure 4a. The mini-gene structure is depicted. Blue arrow: forward primer to detect PCPA; Black arrow: forward primer to detect polyadenylation at 3′ end; Gray arrowhead: 5′ splice site mutation; Green and yellow arrowheads: polyadenylation signals in intron and at 3′ end, respectively. An unspliced and normally cleaved product was too large to detect.
(d) Quantitation of PCPA and normal polyadenylation at 3′ end in control and U1 AMO treated cells was performed using real-time PCR. Error bars indicate s.d. (n=3). PCPA in panel a, b, c and d: premature cleavage and polyadenylation.