Abstract
This study was undertaken to optimize the conditions for the extraction of antibacterial activity of Perilla frutescens var. acuta leaf against Pseudomonas aeruginosa KCTC 2004 using the evolutionary operation-factorial (EVOP) design technique. Increased antibacterial activity was achieved at higher extraction temperatures and with a longer extraction time. Antibacterial activity was not affected by differing ethanol concentration in the extraction solvent. The maximum antibacterial activity of ethanolic extract of P. frutescens var. acuta leaf against P. aeruginosa, determined by the EVOP factorial technique, was obtained at an extraction temperature of 80 °C (R = −0.800**), 26 h (R = −0.731**) extraction time, and 50% (R = −0.075) ethanol concentration. The population of P. aeruginosa also decreased from 6.660 log CFU/mL in the initial set to 4.060 log CFU/mL in the third set. Also, scanning electron microscopy study of the ethanolic extract of P. frutescens var. acuta revealed potential detrimental effects on the morphology of P. aeruginosa.
Keywords: Perilla frutescens var. acuta leaf, EVOP-factorial design technique, antibacterial activity, Pseudomonas aeruginosa KCTC 2004
1. Introduction
With increasing bacterial resistance to antibiotics, there is considerable interest in investigating the antimicrobial effects of plant extracts against a range of bacteria, to develop other classes of safe and natural antimicrobials useful for infection control or for the preservation of food [1]. Pseudomonas aeruginosa (KCTC 2004) is a versatile pathogen associated with a broad spectrum of infections in human beings and the food industry. P. aeruginosa dominates proteinaceous foods stored aerobically at chilled temperatures. In pasteurized milk it originates from post-process contamination [2] and spoils the food products. Pseudomonas sp. is widespread in the environment [3,4] and is often responsible, as an opportunistic bacterium, for very serious episodes of infections. P. aeruginosa is also involved in infections ranging widely in location and severity from soft tissue abscesses, bronchopneumonia, otitis externa and otitis media to chronic pneumopathy or mucoviscidosis [5]. This organism has been shown to possess a remarkable capacity to resist antibiotics, either intrinsically or following acquisition of resistance genes [6].
The main advantage of the evolutionary operation (EVOP) factorial design technique [7–9] is to develop more effective approaches for the optimization of an ‘n’ variable system [10,11], using EVOP methodology including response surface methodology (RSM) derived from orthogonal polynomial fitting techniques [12,13]. The identification and evaluation of natural products with optimized levels of antimicrobial activity for the control of infectious pathogens can be considered as an important international challenge for the food and medicine industries.
The Perilla frutescens L. herb belongs to the family Labiatae. P. frutescens var. acuta is an edible plant frequently used as one of the most popular garnishes and food colorants in China, Japan and Korea. The leaves of P. frutescens var. acuta have been shown to be detoxicant, antitussive, antibiotic, and antipyretic [14,15], and are also utilized as a folk medicine for treating intestinal disorders and allergies, particularly in traditional Chinese medical practice [16]. Although the biological activity of P. frutescens var. acuta is well documented, there is no report available on the optimization of the antibacterial activity of the P. frutescens var. acuta leaf against P. aeruginosa using the EVOP-factorial design technique.
In the present study, therefore, the optimal conditions of extraction temperature, extraction time and ethanol concentration on the antibacterial activity of Perilla frutescens var. acuta leaf were investigated using the EVOP-factorial design to maximize the antibacterial activity of Perilla frutescens var. acuta leaf against P. aeruginosa.
2. Results and Discussion
2.1. Optimization of Antibacterial Activity by EVOP-Factorial System
The experimental conditions used in the first set of experiments, the corresponding antibacterial activities of cycle I and II, their differences and average values are presented in Table 1. The error limits, effects and the change in the mean effect were calculated and the results are shown in Table 2. The extraction temperature, extraction time and ethanol concentration of the central point in first set (E10, E20) were 50 °C, 14 h and 50%, respectively.
Table 1.
Experimental design for the three inducer system and results of Set I.
| Experimental Conditions | E10a | E11 | E12 | E13 | E14 | E20 | E21 | E22 | E23 | E24 |
|---|---|---|---|---|---|---|---|---|---|---|
| Temperature (°C) | 50(0)b | 35(−) | 35(−) | 65(+) | 65(+) | 50(0) | 65(+) | 35(−) | 65(+) | 35(−) |
| Time (h) | 14(0) | 8(−) | 20(+) | 8(−) | 20(+) | 14(0) | 20(+) | 8(−) | 8(−) | 20(+) |
| Ethanol concentration (%) | 50(0) | 25(−) | 75(+) | 75(+) | 25(−) | 50(0) | 75(+) | 75(+) | 25(−) | 25(−) |
| Antibacterial activity (cycle I) (log CFU/mL) | 6.24 | 6.75 | 6.13 | 5.70 | 5.30 | 6.05 | 5.02 | 6.70 | 5.49 | 6.50 |
| Antibacterial activity (cycle II) (log CFU/mL) | 6.10 | 6.57 | 6.28 | 5.52 | 5.07 | 6.28 | 4.89 | 6.49 | 5.70 | 6.36 |
| Difference (cycle I-cycle II) (log CFU/mL) | 0.14 | 0.18 | −0.15 | 0.18 | 0.23 | 0.23 | 0.13 | 0.21 | −0.21 | 0.14 |
| Average activity (log CFU/mL) | 6.170 (a10) | 6.660 (a11) | 6.205 (a12) | 5.610 (a13) | 5.185 (a14) | 6.165 (a20) | 4.955 (a21) | 6.595 (a22) | 5.595 (a23) | 6.430 (a24) |
E10 to E24 = experiments;
Numbers in parentheses are the coded symbols of levels of the extraction conditions.
Table 2.
Calculation worksheet of the effects of the three-variable system and magnitude of effects and error limits of Set I.
| Experimental Conditions | Calculation of Effects | |
|---|---|---|
| Temperature | 1/4(a13 + a14 + a21 + a23 − a11 − a12 − a22 − a24) | −1.1363 |
| Time | 1/4(a12 + a14 + a21 + a24 − a11 − a13 − a22 − a23) | −0.4213 |
| Ethanol concentration | 1/4(a12 + a13 + a21 + a22− a11 − a14 − a23 − a24) | −0.1263 |
| Temperature × time | 1/4(a11 + a14 + a21 + a22 − a12 − a13 − a23 − a24) | −0.1113 |
| Temperature × ethanol concentration | 1/4(a11 + a13 + a21 + a24 − a12 − a14 − a22 − a23) | 0.0188 |
| Time × ethanol concentration | 1/4(a11 + a12 + a21 + a23 − a13 − a14 − a22 − a24) | −0.1013 |
| Temperature × time × ethanol concentration | 1/4(a21 + a22 + a23 + a24 − a11 − a12 − a13 − a14) | −0.0213 |
| Change in mean effect | 1/10(a11 + a12 + a13 + a14 + a21 + a22 + a23 + a24 − 4a10 − 4a20) | −0.2105 |
| Standard deviation (σ) | 1/2(σ1 + σ2)=1/2(R1 × fk,n + R2 × fk,n) (1) | 0.1230 |
| Error limits: | ||
| for average | ±1.414σ (±2σ/√n) | 0.1739 |
| for effects | ±1.004σ (±0.71 × 2σ/√n) | 0.1235 |
| for change in mean | ±0.891σ (±0.63 × 2σ/√n) | 0.1096 |
R1: (largest difference − smallest difference) in block 1; R2: (largest difference − smallest difference) in block 2; fk,n = constant depending on number of replications (n) and number of experiments (k) per cycle = 0.3 for n = 2 and k = 5.
In the first set, the error limits for average, effects and changes in mean were 0.1739, 0.1235 and 0.1096, respectively. The change in mean effect was −0.2105. According to the decision-making procedure, after calculating the change in the mean effect and error limit, an examination was necessary to determine whether any change in the control (search level) experimental conditions would help to improve the objective function [10]. The optimum condition was achieved when the effect was smaller than the error limit while the change in the mean effect was large. Moreover, because the dependent variables are the population of P. aeruginosa in which growth was suppressed by addition of P. frutescens var. acuta leaf extract, the optimum point was reached when the code of mean effect was negative.
The determination of the magnitude of the change in mean effect, which is negative and large, compared to the error limit, is a requirement in order to confirm the achievement of the optimum condition. Such a situation, where some of the effects are larger in comparison to the error limit, does not ensure that the conditions in the search region (E10, E20) of the first set is the actual optimum and a second set of experiments is called for.
In the second set, the search level (E10, E20) was fixed at the best condition of Set I, at a level of E21, in which the number of P. aeruginosa was 4.955 log CFU/mL. The extraction temperature, extraction time and ethanol concentration of the central point in the second set (E10, E20) were 65 °C, 20 h and 75%, respectively. The experimental conditions and the results of the Set II experiments are presented in Table 3 and the effects and error limits are shown in Table 4. In the second set, the error limits for average, effects and changes in mean was 0.1612, 0.1145 and 0.1016, respectively. The change in mean effect was −0.0685. The most effective antibacterial activity (4.095 log CFU/mL) was obtained at E14. The extraction temperature, extraction time and ethanol concentration of the E14 point in the second set were 80 °C, 26 h and 50%, respectively. In this case, not all of the effects were smaller than the error limit, and the change in mean effect was smaller compared to the error limit even though it is positive. It has been reported that if all or any of the effects are larger than the error limits, the change in the experimental conditions may yield better results [10].
Table 3.
Experimental design for three inducer system and results of Set II.
| Experimental Conditions | E10 | E11 | E12 | E13 | E14 | E20 | E21 | E22 | E23 | E24 |
|---|---|---|---|---|---|---|---|---|---|---|
| Temperature (°C) | 65(0) | 50(−) | 50(−) | 80(+) | 80(+) | 65(0) | 80(+) | 50(−) | 80(+) | 50(−) |
| Time (h) | 20(0) | 14(−) | 26(+) | 14(−) | 26(+) | 20(0) | 26(+) | 14(−) | 14(−) | 26(+) |
| Ethanol concentration (%) | 75(0) | 50(−) | 100(+) | 100(+) | 50(−) | 75(0) | 100(+) | 100(+) | 50(−) | 50(−) |
| Antibacterial activity (cycle I) (log CFU/mL) | 4.77 | 6.19 | 4.40 | 4.80 | 4.18 | 4.87 | 4.25 | 6.11 | 4.50 | 4.51 |
| Antibacterial activity (cycle II) (log CFU/mL) | 4.90 | 6.03 | 4.59 | 5.01 | 4.01 | 4.69 | 4.40 | 5.95 | 4.65 | 4.71 |
| Difference (cycle I-cycle II) (log CFU/mL) | −0.13 | 0.16 | −0.19 | −0.21 | 0.17 | 0.18 | −0.15 | 0.16 | −0.15 | −0.20 |
| Average activity (log CFU/mL) | 4.835 (a10) | 6.110 (a11) | 4.495 (a12) | 4.905 (a13) | 4.095 (a14) | 4.780 (a20) | 4.325 (a21) | 6.030 (a22) | 4.575 (a23) | 4.610 (a24) |
E10 to E24 = experiments;
Numbers in parentheses are the coded symbols of levels of the extraction conditions.
Table 4.
Calculation worksheet of the effects of the three-variable system and magnitude of effects and error limits of Set II.
| Experimental Conditions | Calculation of Effects |
|---|---|
| Temperature | −0.8363 |
| Time | −1.0238 |
| Ethanol concentration | 0.0913 |
| Temperature × time | 0.4938 |
| Temperature × ethanol concentration | 0.1888 |
| Time × ethanol concentration | −0.0338 |
| Temperature × time × ethanol concentration | −0.0163 |
| Change in mean effect | 0.0685 |
| Standard deviation (σ) | 0.1140 |
| Error limits: | |
| for average | 0.1612 |
| for effects | 0.1145 |
| for change in mean | 0.1016 |
R1: (largest difference − smallest difference) in block 1; R2: (largest difference − smallest difference) in block 2; fk,n = constant depending on number of replications (n) and number of experiments (k) per cycle = 0.3 for n = 2 and k = 5.
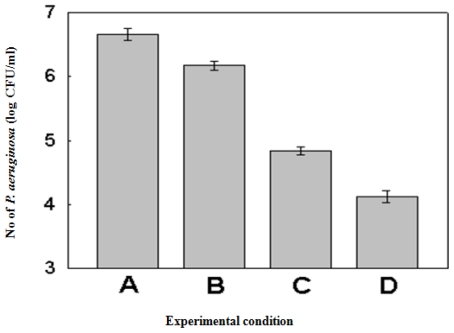
From the above situation, the third set of experiments was designed in which the best condition of Set II (E14) was selected as the search level (E10, E20) for Set III. The experimental conditions and the results of Set III are shown in Table 5 and the calculated effects and error limits are presented in Table 6. In the EVOP-factorial design, the effects remain smaller than the error limits while the changes in the mean effect remain larger and positive so as to reach the optimum level. Thus, in the experiments of third set, we were able to arrive at the proper optimum condition, in which all effects were smaller than the error limit and the change in mean effect is large and positive. As shown in Figure 1, the population of P. aeruginosa decreased from 6.660 log CFU/mL in the initial set to 4.060 log CFU/mL in the third set.
Table 5.
Experimental design for the three inducer system and results of Set III.
| Experimental Conditions | E10 | E11 | E12 | E13 | E14 | E20 | E21 | E22 | E23 | E24 |
|---|---|---|---|---|---|---|---|---|---|---|
| Temperature (°C) | 80(0) | 65(−) | 65(−) | 95(+) | 95(+) | 80(0) | 95(+) | 65(−) | 95(+) | 65(−) |
| Time (h) | 26(0) | 20(−) | 32(+) | 20(−) | 32(+) | 26(0) | 32(+) | 20(−) | 20(−) | 32(+) |
| Ethanol concentration (%) | 50(0) | 25(−) | 75(+) | 75(+) | 25(−) | 50(0) | 75(+) | 75(+) | 25(−) | 25(−) |
| Antibacterial activity (cycle I) (log CFU/mL) | 4.21 | 4.38 | 4.51 | 4.41 | 4.4. | 3.96 | 4.24 | 4.65 | 4.50 | 4.39 |
| Antibacterial activity (cycle II) (log CFU/mL) | 4.03 | 4.57 | 4.39 | 4.50 | 4.20 | 4.16 | 4.35 | 4.44 | 4.37 | 4.49 |
| Difference (cycle I-cycle II) (log CFU/mL) | 0.18 | −0.15 | 0.12 | −0.09 | 0.20 | −0.20 | −0.11 | 0.21 | 0.13 | −0.10 |
| Average activity (log CFU/mL) | 4.120 (a10) | 4.475 (a11) | 4.450 (a12) | 4.455 (a13) | 4.300 (a14) | 4.060 (a20) | 4.295 (a21) | 4.545 (a22) | 4.435 (a23) | 4.440 (a24) |
E10 to E24 = experiments;
Numbers in parentheses are the coded symbols of levels of the extraction conditions.
Table 6.
Calculation worksheet of the effects of the three-variable system and magnitude of effects and error limits of Set III.
| Experimental Conditions | Calculation of Effects |
|---|---|
| Temperature | −0.1063 |
| Time | −0.1063 |
| Ethanol concentration | 0.0238 |
| Temperature × time | −0.0413 |
| Temperature × ethanol concentration | −0.0163 |
| Time × ethanol concentration | −0.0213 |
| Temperature × time × ethanol concentration | 0.0088 |
| Change in mean effect | 0.2675 |
| Standard deviation (σ) | 0.1140 |
| Error limits: | |
| for average | 0.1612 |
| for effects | 0.1145 |
| for change in mean | 0.1016 |
R1: (largest difference − smallest difference) in block 1; R2: (largest difference − smallest difference) in block 2; fk,n = constant depending on number of replications (n) and number of experiments (k) per cycle = 0.3 for n = 2 and k = 5.
Figure 1.
Comparison of the antibacterial activity of Perilla frutescens var. acuta leaf against P. aeruginosa at the central point of each set. (A) E11 of Set I (extraction temperature; 35 °C, extraction time; 8 h, ethanol concentration; 25%); (B) central point of Set I (Extraction temperature; 50 °C, extraction time; 14 h, ethanol concentration; 50%); (C) central point of Set II (Extraction temperature; 65 °C, extraction time; 20 h, ethanol concentration; 75%); (D) central point of Set III (Extraction temperature; 80 °C, extraction time; 26 h, ethanol concentration; 50%).
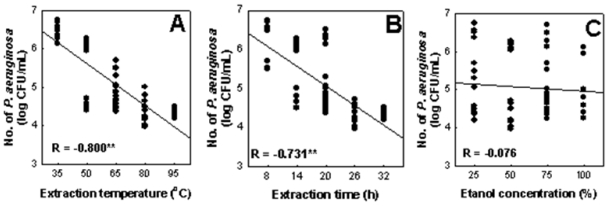
In this study, it was shown that higher antibacterial activity was achieved at a higher extraction temperature of 80 °C (R = −0.800**) and in a longer extraction time of 26 h (R = −0.731**). However, antibacterial activity of P. frutescens var. acuta leaf extract against P. aeruginosa was not affected by the presence of different ethanol concentrations in the extraction solvent (R = −0.076), as shown in Figure 2. Therefore, the maximum antibacterial activity of P. frutescens var. acuta leaf against P. aeruginosa, determined by the EVOP-factorial technique, was obtained at 80 °C extraction temperature, 26 h extraction time and 50% ethanol concentration. These conditions were also found in our previous study conducted on Coptidis rhizoma against Streptococcus mutans [17].
Figure 2.
Plots of the responses against the three independent variables in EVOP. ** p < 0.01. (A) Extraction temperature; (B) extraction time; (C) ethanol concentration.
Although the optimal condition of P. frutescens var. acuta on antibacterial activity is well described in this study, the extraction yield and bioactive compounds in P. frutescens var. acuta are highly variable depending on environmental conditions. Therefore, further studies are needed to compare the effects of various seasonal samples of P. frutescens var. acuta leaf on antibacterial activity using EVOP-factorial design technique.
2.2. Scanning Electron Microscopy (SEM)
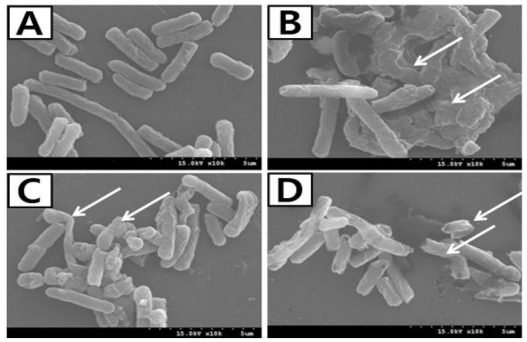
Further, a SEM study of was carried out to visualize the effects of the ethanolic extract of P. frutescens var. acuta leaf on the morphology of P. aeruginosa and demonstrated altered cell morphology as compared to a control group (Figure 3). Control cells in the absence of the extract showed a regular, smooth surface (Figure 3A). In contrast, cells inoculated with the ethanolic extract of P. frutescens var. acuta leaf revealed severe detrimental effects on the morphology of the cell membrane, showing disruption and lysis of the membrane integrity (Figure 3B). Exposure of P. frutescens leaf extract to P. aeruginosa revealed extensive surface collapse and wrinkled abnormalities on the morphology of the cells along with some small clefts formation (Figure 3C, 3D) and these finding are in strong agreement with a previous report [16]. Besides, several researchers have reported the effects of various plant extracts on the morphology of pathogenic bacteria including P. aeruginosa [18,19].
Figure 3.
Scanning electron micrographs of P. aeruginosa incubated with the optimum concentration of ethanolic leaf extract of Perilla frutescens var. acuta (A) Control (absence of extract); (B) disruption and lysis of membrane integrity; (C) wrinkled abnormalities and cleft formation; (D) abnormal breaking of cell.
3. Experimental Section
3.1. Plant Material
The leaves of P. frutescens var. acuta were obtained from Yakrung market, Daegu, Republic of Korea, in July 2008. The specimen was lyophilized for 48–72 h after storage at −70 °C. Freeze dried samples were pulverized with a blender (HJM-7000, Hanil, Korea). Extra pure grade ethanol solvent was purchased from Daemyung Scientific Co., Daegu, Republic of Korea. Chemical reagents were obtained from Sigma Co. (St. Louis, MO, USA), unless otherwise stated.
3.2. Microorganism
Pseudomonas aeruginosa KCTC 2004 was used in the antibacterial assay. The strain was obtained from the Korea Food and Drug Administration (KFDA), Daegu, Republic of Korea. Active cultures for experimental use were prepared by transferring a loopful of cells from stock cultures to flasks and inoculatino in Luria-Bertani (LB) broth medium at 37 °C for 24 h. A culture of the bacterial strain was maintained on LB agar medium at 4 °C.
3.3. Preparation of Extracts from Various Conditions
To design an experiment for establishing proper extraction conditions, 20 g of sample was hydrolyzed and extracted in a reflux extraction apparatus under different extraction temperatures (35, 50, 65, 80 and 95 °C), extraction times (8, 14, 20, 26 and 32 h) and ethanol concentrations (25, 50, 75 and 100%) under EVOP factorial design and then freeze dried in order to establish the proper extraction conditions. The concentration of P. frutescens var. acuta extract used for the determination of the microbial population was 0.1%. Samples were homogenized for 30 seconds and serially diluted with peptone water (Difco, USA), as needed for the determination of microbial populations.
3.4. EVOP-Factorial Design Technique
In this study, the optimization of antibacterial activity of Perilla frutescens var. acuta leaf was investigated using the EVOP-factorial design technique, which was first proposed for the optimization of different operational parameters in a functioning chemical processing plant. The main advantage of EVOP methodology is its clear-cut decision making procedure, which indicates the change in influencing variables toward the desired maxima or minima [20]. The EVOP-factorial design technique was applied to select the optimum conditions of three extraction factors (temperature, time, and ethanol concentration) in different experiments [9]. Firstly, the control or search level experimental conditions (E10, E20) were selected based on the results of earlier investigation on the effect of individual extraction conditions on the antibacterial activity of the ethanolic extract of P. frutescens var. acuta leaf [21]. Secondly, the new experimental conditions (Ebe) were selected with lower and higher levels of inducers compared to the initial search level (Eb0). Antibacterial activities of P. frutescens var. acuta leaf extract were estimated following the given assay procedure and recorded for cycle I and II. Differences in the antibacterial activities between cycle I and II, and average antibacterial activities were calculated to estimate the effects and error limits. The magnitudes of effects, error limits and change in mean effect were examined as per the decision making procedure to arrive at the optimum level. When the experimental results of the first set did not reach to the satisfactory level of optimum conditions, a second set of experiments was planned, selecting the best condition of the first set as the new search level for the second set. This procedure was repeated until the optimum condition was obtained.
Further, to determine the antibacterial activity of P. frutescens var. acuta leaf extract, enumeration of viable counts on LB plates was monitored as follows: 1 mL of the re-suspended culture was diluted into 9 mL buffer peptone water, thereby diluting it by 10-fold. In total, 0.1 mL sample of each treatment was diluted and spread on the surface of LB agar. The colonies were counted after 24 h of incubation at 37 °C.
3.5. Scanning Electron Microscopic (SEM) Analysis
To determine the efficacy of the extract of P. frutescens var. acuta leaf on the morphology of P. aeruginosa, SEM studies were performed using the optimum extraction conditions of the ethanolic extract of P. frutescens var. acuta leaf. Controls were prepared without extract. Further, to observe the morphological changes, the method of SEM was modified from Kockro et al.’s method [22]. The bacterial sample was washed gently with 50 mM/L phosphate buffer solution (pH 7.2), fixed with 2.5 g 100/mL glutaraldehyde and 1 g/100 mL osmic acid solution. The specimen was dehydrated using sequential exposure to ethanol concentrations ranging from 30–100%. The ethanol was replaced by tertiary butyl alcohol. After dehydration, the specimen was dried with CO2. Finally, the specimen was sputter-coated with gold in an ion coater for 2 min, followed by microscopic examinations (S-4300; Hitachi, Japan).
4. Conclusions
Overall, our results demonstrate that the application of EVOP-factorial design technique could serve as a potential tool to determine the optimum extraction conditions required to achieve the desired levels of antibacterial activity of natural products and their extracts for their potential utilization in the food industry to control food-borne pathogenic bacteria.
References
- 1.Careaga M, Fernandez E, Dorantes L, Mota L, Jaramillo ME, Hernandez-Sanchez H. Antibacterial activity of Capsicum extract against Salmonella typhimurium and Pseudomonas aeruginosa inoculated in raw beef meat. Int. J. Food Microbiol. 2003;83:331–335. doi: 10.1016/s0168-1605(02)00382-3. [DOI] [PubMed] [Google Scholar]
- 2.Eneroth A, Ahrne S, Molin G. Contamination routes of Gram-negative spoilage bacteria in the production of pasteurised milk, evaluated by randomly amplified polymorphic DNA (RAPD) Int. Dairy J. 2000;10:325–331. [Google Scholar]
- 3.Ringen LM, Drake CH. A study of the incidence of Pseudomonas aeruginosa from various natural sources. J. Bacteriol. 1952;64:841–845. doi: 10.1128/jb.64.6.841-845.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheater DWF, Mara DD, Jawad L, Oragui J. Pseudomonas aeruginosa and Escherichia coli in sewage and fresh water. Water Res. 1980;14:713–721. [Google Scholar]
- 5.Molina DN, Colon M, Bermudez RH, Ramirez-Ronda CH. Unusual presentation of Pseudomonas aeruginosa infections: a review. Biol. Assoc. Med. 1991;83:160–163. [PubMed] [Google Scholar]
- 6.Mesaros N, Nordmann P, Plesiat P. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin. Microbiol. Inf. 2007;13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 7.Box GEP. Evolutionary operation: a method of increasing industrial productivity. Appl. Stat. 1957;6:81–101. [Google Scholar]
- 8.Kim SJ, Park SY, Kim CW. A Novel Approach to the Production of Hyaluronic Acid by Streptococcus zooepidemicus. J. Microbiol. Biotechnol. 2006;16:1849–1855. [Google Scholar]
- 9.Mukherjee G, Banerjee R. Evolutionary operation-factorial design technique for optimization of conversion of mixed agroproducts into gallic acid. Appl. Biochem. Biotechnol. 2004;118:33–46. doi: 10.1385/abab:118:1-3:033. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee R, Bhattacharyya BC. Evolutionary operation (EVOP) to optimize three-dimensional biological experiments. Biotechnol. Bioeng. 1993;41:67–71. doi: 10.1002/bit.260410109. [DOI] [PubMed] [Google Scholar]
- 11.Tunga R, Banerjee R, Bhattacharyya BC. Optimization of n variable biological experiments by evolutionary operation-factorial design technique. J. Biosci. Bioeng. 1999;87:224–230. doi: 10.1016/s1389-1723(99)89017-3. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan K, Trivedi U, Patel KC. Application of response surface methodology for optimization of lactic acid production using date juice. J. Microbiol. Biotechnol. 2006;16:1410–1415. [Google Scholar]
- 13.Kvist T, Thyregod P. Using evolutionary operation to improve yield in biotechnological processes. Qual. Reliab. Eng. Int. 2005;21:457–463. [Google Scholar]
- 14.Liu JH, Steigel A, Reininger E, Bauer R. Two new prenylated 3-benzoxepin derivatives ascyclooxygenase inhibitors from Perilla frutescens var. acuta. J. Nat. Prod. 2000;63:403–405. doi: 10.1021/np990362o. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura Y, Ohto Y, Murakami A, Ohigashi H. Superoxide scavenging activity of rosmarinic acid from Perilla frutescens Britton var. acuta f. Viridis. J. Agric. Food Chem. 1998;46:4545–4550. [Google Scholar]
- 16.Nakazawa T, Ohsawa K. Metabolites of orally administered Perilla frutescens extract in rats and humans. Biol. Pharm. Bull. 2000;23:122–127. doi: 10.1248/bpb.23.122. [DOI] [PubMed] [Google Scholar]
- 17.Choi UK, Kim MH, Lee NH. Optimization of antibacterial activity by gold-thread (Coptidis Rhizoma Franch) against Streptococcus mutans using evolutionary operation-factorial design technique. J. Microbiol. Biotechnol. 2007;17:1880–1884. [PubMed] [Google Scholar]
- 18.Benli M, Kaya I, Yigit N. Screening antimicrobial activity of various extracts of Artemisia dracunculus L. Cell Biochem. Funct. 2007;25:681–686. doi: 10.1002/cbf.1373. [DOI] [PubMed] [Google Scholar]
- 19.Eloff JN. Antibacterial activity of Marula (Sclerocarya birrea (A. rich.) Hochst. subsp. caffra (Sond.) Kokwaro) (Anacardiaceae) bark and leaves. J. Ethnopharmacol. 2001;76:305–308. doi: 10.1016/s0378-8741(01)00260-4. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee G, Banerjee R. Evolutionary operation-factorial design technique for optimization of conversion of mixed agroproducts into gallic acid. Appl. Biochem. Biotechnol. 2004;118:33–46. doi: 10.1385/abab:118:1-3:033. [DOI] [PubMed] [Google Scholar]
- 21.Kim MH, Lee NH, Lee MH, Kwon DJ, Choi UK. Antimicrobial activity of aqueous ethanol extracts of Phenilla frutescens var. acuta leaf. Korean J. Food Culture. 2007;22:266–273. [Google Scholar]
- 22.Kockro RA, Hampl JA, Jansen B, Peters G, Scheihing M, Giacomelli R. Use of scanning electron microscopy to investigate the prophylactic efficacy of rifampin-impregnated CSF shunt catheters. J. Med. Microbiol. 2000;49:441–450. doi: 10.1099/0022-1317-49-5-441. [DOI] [PubMed] [Google Scholar]