Table 4.
The bear genes showing rapid evolution in bear lineage (CDS coverage ≥ 80%).
| Gene Symbol | P valuea | 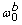 |
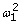 |
CDS Coverage d | Biological functionse |
|---|---|---|---|---|---|
| PNPLA4 | 0.001 | 0.145 | 0.649 | 0.984 | Lipid catabolic process |
| CDC42SE1 | 0.001 | 0.0001 | 0.433 | 0.987 | Signal transduction; Regulation of cell shape |
| CNPY2 | 0.002 | 0.027 | 0.402 | 1 | Unknown |
| TMED2 | 0.011 | 0.006 | 0.211 | 1 | Vesicle-mediated transport; |
| CSRP3 | 0.012 | 0.014 | 0.113 | 0.994 | Regulation of the force of heart contraction; Cellular calcium ion homeostasis; |
| CHCHD1 | 0.016 | 0.144 | 4.156 | 0.991 | Unknown |
| TNNI3 | 0.017 | 0.018 | 0.145 | 0.86 | Cardiac muscle contraction; Regulation of systemic arterial blood pressure by ischemic conditions; Cellular calcium ion homeostasis; |
| APOHL | 0.018 | 0.226 | 0.506 | 0.962 | Negative regulation of endothelial cell proliferation; Triacylglycerol metabolic process; Blood coagulation, intrinsic pathway; |
| C1orf52 | 0.019 | 0.078 | 0.711 | 1 | Unknown |
| PLNM | 0.02 | 0.058 | Inf* | 0.981 | Regulation of the force of heart contraction; Cellular calcium ion homeostasis; Blood circulation |
| C19orf39 | 0.021 | 0.301 | 0.948 | 0.925 | Unknown |
| BPHL | 0.024 | 0.127 | 0.314 | 0.859 | Proteolysis; Response to toxin; Cellular amino acid and derivative metabolic process |
| YIPF3 | 0.028 | 0.021 | 0.08 | 0.854 | Cell differentiation |
| DR1 | 0.029 | 0.001 | 0.191 | 0.824 | Negative regulation of transcription |
| RBM8AM | 0.033 | 0.0001 | 0.068 | 0.821 | RNA processing; mRNA binding |
| POLR2C | 0.039 | 0.003 | 0.037 | 0.876 | Transcription initiation |
| ATP6V1FH | 0.042 | 0.009 | Inf* | 0.991 | Ion transport; ATP synthesis coupled proton transport; |
| CISD2 | 0.043 | 0.011 | 0.15 | 1 | Regulation of cellular respiration |
ap-value calculated from the LRT under chi-squared distribution with df = 1.
b ω1 is the Ka/Ks value of the bear lineage; c ω0 is the average Ka/Ks value in the rest of lineages.
dThe coverage of protein coding region of bear EST using human RefSeq sequence as reference.
eBiological functions obtained from GO annotations.
* In these cases, the rate of synonymous substitution reaches zero (Ks = 0) and Ka/Ks diverges and represented as Inf here.
H, L, MThe gene shows significantly differential expression (P < 0.05, one-way ANOVA) in heart, liver, or skeletal muscle tissue respectively.
