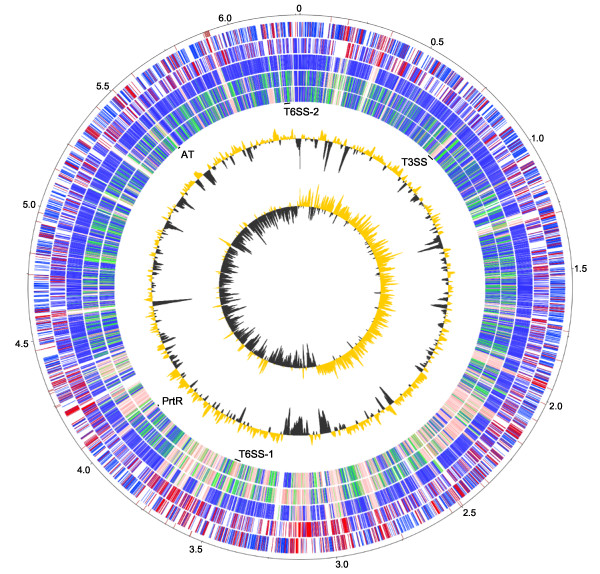
Figure 1.
Circular representation of the improved, high-quality draft genome sequence of WH6. The outer scale designates the coordinates in half million base pair increments. The red ticks indicate physical gaps. Circles 2 and 3 show the predicted coding regions of WH6 on the positive and negative strands, respectively. Coding regions are colored to highlight orthologous (blue) and 1567 unique (red) coding regions of WH6 relative to the other sequenced P. fluorescens. Circles 4, 5, and 6 show orthologs (BLASTP e-value ≤ 1 × 10-7) of SBW25, Pf-5, and Pf0-1, respectively. The extent of homology relative to WH6 is depicted using a heat map of arbitrarily chosen bins; dark blue: orthologs with greater than 80% homology over the length of the gene; green: orthologs with between 60-80% homology over the length of the gene; pink: orthologs with between 20-60% homology over the length of the gene; white: no homology (less than 20% homology over the length of the gene). The positions of loci of interest are also denoted (see corresponding text for more details). Circles 7 and 8 show GC% (gold >60.6% average; gray < 60.6% average) and GC-skew.