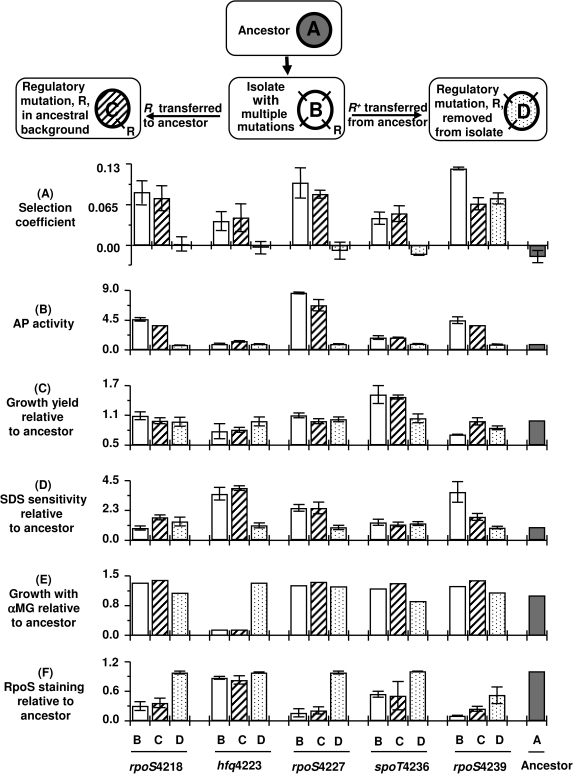
FIG. 3.—
Fitness and phenotypes in strains with altered regulatory genes. We compared the five evolved isolates (Type B in the schematic) and two classes of derivative strains manipulated for each regulatory allele by cotransduction with linked markers (see Materials and Methods). In one class, each mutated regulatory gene (R) was transferred into a clean ancestral background (Type C). In the other class, the regulatory mutation present in each isolate was replaced by the ancestral regulatory gene (Type D) while retaining the other evolved mutations. (A) Competitive fitness was measured for Type B, C, and D strains in Pi-limited chemostats. Competitions were all against a reference strain BW3454 containing a metC::Tn10 countable marker; this strain is marginally fitter than ancestor. Competing strains were mixed 50:50 after 16 h individual acclimatization in chemostats. Selection coefficients (Dykhuizen and Hartl 1983) were calculated from changes in population proportions. The mean and standard deviations were obtained from 3 to 5 replicates. (B) The alkaline phosphatase was measured using ONP-Pi as substrate after growth in media containing 30 μM KH2PO4. (C) The growth yield of strains was measured from the absorbance in L-broth after overnight growth to steady state and shown relative to ancestor. (D) Sensitivity to 3% SDS was measured by growth in microtitre plates, with densities divided by that of the ancestor shown. (E) Sensitivity to 1% methyl α-glucoside (α-MG) was measured in patches on glycerol plates. Patches were scanned densitometrically, and density relative to ancestor is shown. (F) The RpoS level was estimated by glycogen staining as in figure 2 and scanning densities relative to that of ancestor.