Abstract
Evolutionary biologists are increasingly comparing gene expression patterns across species. Due to the way in which expression assays are normalized, such studies provide no direct information about expression per gene copy (dosage responses) or per cell and can give a misleading picture of genes that are differentially expressed. We describe an assay for estimating relative expression per cell. When used in conjunction with transcript profiling data, it is possible to compare the sizes of whole transcriptomes, which in turn makes it possible to compare expression per cell for each gene in the transcript profiling data set. We applied this approach, using quantitative reverse transcriptase-polymerase chain reaction and high throughput RNA sequencing, to a recently formed allopolyploid and showed that its leaf transcriptome was approximately 1.4-fold larger than either progenitor transcriptome (70% of the sum of the progenitor transcriptomes). In contrast, the allopolyploid genome is 94.3% as large as the sum of its progenitor genomes and retains ≥93.5% of the sum of its progenitor gene complements. Thus, “transcriptome downsizing” is greater than genome downsizing. Using this transcriptome size estimate, we inferred dosage responses for several thousand genes and showed that the majority exhibit partial dosage compensation. Homoeologue silencing is nonrandomly distributed across dosage responses, with genes showing extreme responses in either direction significantly more likely to have a silent homoeologue. This experimental approach will add value to transcript profiling experiments involving interspecies and interploidy comparisons by converting expression per transcriptome to expression per genome, eliminating the need for assumptions about transcriptome size.
Keywords: transcriptome size, transcriptome-normalized expression, genome-normalized expression, genome doubling, gene dosage responses
Introduction
A growing number of transcript profiling studies, primarily using microarrays, have compared global expression patterns among closely related species, providing insights into a range of important evolutionary questions. Included among these are studies characterizing the selection pressures acting on gene expression in primates (Enard et al. 2002; Gilad et al. 2006), studies quantifying gene expression variation within and between populations or species of teleost fishes (Oleksiak et al. 2002), fruit flies (Rifkin et al. 2003), fungi (Andersen et al. 2008), and plants (Hammond et al. 2006), and several studies examining the effects of hybridization and genome doubling on gene expression in plants (Hegarty et al. 2005, 2006, 2008; Udall et al. 2006; Wang et al. 2006a; Flagel et al. 2008; Hovav et al. 2008a, 2008b; Rapp et al. 2009). The advent of next generation sequencing technologies is likely to accelerate further the increase in such studies by removing many of the challenges associated with microarrays for interspecies comparisons (Gilad and Borevitz 2006; Blencowe et al. 2009; Gilad et al. 2009; Rokas and Abbot 2009).
Transcript profiling studies provide information about the relative abundances of transcripts. These and other expression assays such as reverse transcriptase-polymerase chain reaction (RT-PCR) and RNA blots require normalization to correct for differences in amount of RNA template, as well as for other technical biases (Thellin et al. 1999; Quackenbush 2002), before comparisons can be made between samples. One or a few housekeeping genes are typically used as loading controls for RNA blots and RT-PCR assays, on the assumption that these genes are stably expressed across samples, thereby indicating the total amount of RNA used. With microarrays, raw data are generally normalized to total signal intensity (Quackenbush 2002) on the assumption that if the features on the array are a complete or unbiased sampling of the transcriptome, total signal intensity is a reasonable proxy for the whole transcriptome. For RNA sequencing (RNA-Seq) data, read counts per gene are typically divided by gene length and total read count per sample (expressed as reads per kilobase per million [RPKM]) to achieve comparable normalization (Marioni et al. 2008; Mortazavi et al. 2008). Consequently, for each of these assays, apparent differences in the expression of a gene between two samples are actually differences in expression per unit of RNA or “per transcriptome” (Kanno et al. 2006).
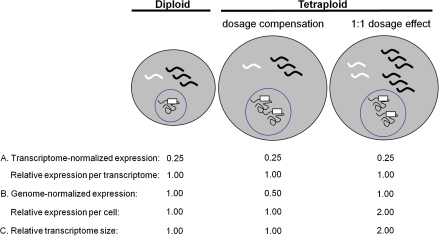
Without information about the sizes of the two transcriptomes being compared, no inferences can be drawn from transcriptome-normalized expression about expression per gene copy or expression per cell (fig. 1). Any difference in expression per cell between two samples that is proportional to the change in total transcriptome size will appear as equal expression per transcriptome. For example, in comparing a tetraploid with a diploid progenitor, genes showing equal expression per transcriptome (combining expression from the two homoeologous copies in the case of the tetraploid; fig. 1) could have equal numbers of transcripts per cell (if the transcriptomes are of equal size), or there could be twice as many transcripts per cell in the polyploid (if the polyploid transcriptome is doubled in size relative to the diploid; fig. 1). Conversely, genes exhibiting repression in the polyploid on a per transcriptome basis could be expressed at an equal or even greater level per cell, again depending on the relative sizes of the two transcriptomes.
FIG. 1.—
A comparison of transcriptome-normalized expression data versus genome-normalized expression data. Gray circles represent cells, and wavy lines represent transcripts, with the diploid cell having a total of four transcripts in its transcriptome. Black circles represent nuclei, squiggly lines represent gDNA, and white boxes represent the genes encoding the white transcripts. (A) Transcriptome-normalized expression. Expression of the white transcript, measured on a per transcriptome basis, is 0.25 (1 transcript out of a total of 4 transcripts) in the diploid. The same transcriptome-normalized expression values are obtained in two tetraploids showing different expression levels per cell, illustrating that transcriptome-normalized measurements do not provide information on transcript abundance per genome (dosage response), or per cell. (B) Genome-normalized expression. If the expression of the white transcript is instead normalized to genome copy number (1 for the diploid, 2 for the tetraploid), differences in transcript abundance per cell become apparent, and dosage responses can be determined. Relative expression per cell in the tetraploid is simply two times the genome-normalized expression. (C) Relative transcriptome size. Tetraploid transcriptome size (relative to the diploid transcriptome) can then be estimated by dividing relative expression per cell by relative expression per transcriptome.
The unstated assumption of expression studies is that the transcriptomes being compared are of equal size. This seems an unwarranted assumption, particularly when comparing polyploids and diploids, because transcriptome sizes are likely to differ due to genome-wide differences in gene dosage. But even for comparisons not involving ploidy differences, the potential exists for transcriptome sizes to differ, especially when comparisons are made across tissue types, developmental stages, or species, for which microarray experiments frequently observe dramatic differences in transcriptome-normalized expression profiles (Hammond et al. 2006; Andersen et al. 2008). Given numerous differences in transcriptome-normalized expression, what is the net effect on transcriptome size? In the absence of a method to quantify this effect, it is not possible to determine what such differences, at the level of individual genes, mean in terms of transcript abundance per cell. Thus, a method to estimate relative transcriptome sizes is needed for determining expression differences per cell based on transcriptome-normalized expression profiling data. This is particularly true for expression studies of polyploids.
Most, if not all, flowering plants have experienced one or more whole genome duplications (polyploidy events) during their evolutionary histories (Cui et al. 2006; Tang et al. 2008), and an estimated 15% of angiosperm speciation events are associated with increases in ploidy (Wood et al. 2009). Polyploids often appear to be more successful than their diploid progenitors, as measured by broader geographical ranges (Ehrendorfer 1980; Otto and Whitton 2000), and greater capacity to tolerate stressful environments (Stebbins 1971; Lewis 1980; Grant 1981; Otto and Whitton 2000; Hegarty and Hiscock 2008), and it has been proposed that polyploidy contributed to the survival of several plant lineages through the Cretaceous–Tertiary mass extinction (Fawcett et al. 2009).
Changes in gene expression, due to epigenetic mechanisms, transposon activation, sequence changes, novel combinations of regulatory factors and/or increased gene dosage, are thought to underlie this apparent success (Chen 2007). Consequently, a central focus of polyploidy research is in understanding transcriptional responses to genome duplication.
For every gene duplicated by polyploidy, a range of dosage responses (changes in expression associated with changes in gene dosage) is possible. The two most obvious are dosage compensation, in which expression is modulated to 1.0× diploid levels per cell or 0.5× per genome, and 1:1 dosage effects, resulting in 2.0× diploid expression per cell or 1.0× per genome. Other responses are also possible, including partial dosage compensation (expression between 1.0 and 2.0× diploid level per cell or 0.5 and 1.0× per genome), negative dosage effects (expression <1.0× diploid level per cell or <0.5× per genome), and >1:1 dosage effects (expression >2.0× diploid level per cell or >1.0× per genome). “Dosage effect” and “dosage compensation” refer most clearly to comparisons of an artificial autopolyploid with the diploid genotype from which it was synthesized. In an allopolyploid that combines two differentiated diploid genomes, the situation is more complex. Additivity of the two parental expression levels for a given gene would be the equivalent of a 1:1 dosage effect, with midparent expression levels being analogous to dosage compensation. Regardless of the type of polyploidy involved, the cumulative effect of these dosage responses will dictate to what extent the polyploid transcriptome differs in size from its diploid progenitor transcriptomes.
There is little information available about gene dosage responses following polyploid duplication. In a seminal investigation of a synthetic maize (Zea mays) autopolyploid series, Guo et al. (1996) established that rRNA exhibits a 1:1 dosage effect in response to changes in ploidy, then used rRNA as a loading control for northern blots in order to determine dosage responses for 18 genes. Most of the 18 genes investigated exhibited a 1:1 dosage effect. There were, however, several exceptions, with some genes showing negative dosage effects, others showing >1:1 dosage effects, and others showing variable responses depending on the specific ploidy level (“odd/even effects”). Beyond this study, the literature is largely silent, with no equivalent data available for natural autopolyploids or for natural or synthetic allopolyploids. Thus, it remains an open question how the responses observed by Guo et al. (1996) extend to other genes, other tissues and other species, and how the responses of individual genes sum over the transcriptome as a whole. There exists no literature on overall transcriptome size in polyploids relative to their diploid progenitors.
Here, we have calculated the relative size of an allopolyploid leaf transcriptome by combining genome-normalized expression estimates from a novel quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) assay with transcriptome-normalized expression estimates from RNA-seq. By this approach, we made seven independent measurements of relative transcriptome size, which we then used to test two hypotheses: 1) the allopolyploid transcriptome is equal in size to the midparent transcriptome (genome-wide dosage compensation) and 2) the tetraploid transcriptome is equal to the sum of its progenitor transcriptomes (a genome-wide dosage effects). We then used our estimate of transcriptome size to estimate expression per genome and per cell in the allopolyploid relative to its diploid progenitors, for approximately 15,000 genes in the RNA-Seq data set. This made it possible to quantify the frequency distributions, as well as patterns of homoeologue deployment, for each kind of dosage response.
Materials and Methods
Plant Material
The study group consisted of the natural allopolyploid, Glycine dolichocarpa (2n = 80; designated “T2”) and its diploid progenitors, G. tomentella (2n = 40; “D3”) and G. syndetika (2n = 40; “D4”). (Doyle et al. 2004; Pfeil et al. 2006). The two diploid species, D3 and D4, diverged approximately 2.5 Ma and hybridized to give rise to T2 within the last 100,000 years (Doyle et al. 2004). T2 is therefore a fixed hybrid, whose genome comprises two homoeologous subgenomes, one contributed by D3 and the other by D4. Therefore, at each locus in T2, there is a D3 and a D4 allele, except in cases where the D3 or D4 homoeologue has been lost during the relatively short time since the formation of T2.
Plants were grown in a common growth chamber with a 12:12 h light:dark cycle and 125 μmol/m2 s light intensity. Young, fully expanded leaflets were collected 1.5–2.0 h into the light period and frozen in liquid nitrogen.
Genome-Normalized Expression Assay
In order to estimate relative expression level per genome, we devised a qRT-PCR assay that normalizes cDNA amplification to genomic DNA (gDNA) amplification. The key to this assay is simultaneously extracting both RNA and gDNA from the same tissue so that in vivo RNA/gDNA ratios are preserved. Primers that specifically amplify either cDNA or gDNA were then used for qRT-PCR, allowing for normalization of gene expression (cDNA amplification) to genome copy number (gDNA amplification). This contrasts with typical qRT-PCR assays, in which target cDNA amplification of a target gene is normalized to cDNA amplification of a reference gene.
Leaflets were pooled from six individuals for each biological replicate. Three biological replicates were analyzed per species. RNA and gDNA (total nucleic acid [TNA]) were coextracted from each biological replicate using the BioChain Dr. P Isolation Kit, with the following modifications: 1) centrifugation steps were performed at room temperature. 2) The DNA/RNA pellet obtained from the isopropanol precipitation was washed 3× with 70% EtOH, then resuspended in DEPC H2O/0.1% ethylenediaminetetraacetic acid. This TNA suspension was then used as the template for reverse transcription. RNA, in a mixture with gDNA (∼1 μg TNA), was reverse transcribed with random decamers using the Ambion Retroscript kit.
Primers were designed to be specific to either cDNA or gDNA as follows. For cDNA-specific primers, one or both primers in a pair were designed to span exon–exon splice junctions so that they would not anneal to unspliced gDNA. For gDNA-specific primers, one or both primers were designed to prime at least partially within an intron so that they would not anneal to spliced cDNA. Template specificity was confirmed for all primer pairs by semiquantitative PCR with cDNA and gDNA templates. Primer target sequences were confirmed for each gene in all three species by Sanger sequencing. Primers specific to cDNA were designed for seven genes or gene families (table 1 and supplementary table S1, Supplementary Material online). Primers specific to gDNA were designed to three genes or gene families (supplementary table S1, Supplementary Material online).
Table 1.
Genes and Gene Families for Which Expression Was Analyzed by Genome-Normalized qRT-PCR
| Glycine max Gene IDsa | Annotationb | Unique RPM—T2 | Transcriptome-Normalized Expression Ratio |
|
| T2/D3 | T2/D4 | |||
| Glyma13g23150, Glyma17g11720 | MGD | 30.6 | 2.5 | 1.4 |
| Glyma15g32540 | EMB1473 | 271.6 | 1.1 | 0.7 |
| Glyma04g39380, Glyma06g15520 | Actin | 425.3 | 1.1 | 1.4 |
| Glyma18g03440, Glyma11g34900 | SBPase | 1,260.4 | 1.0 | 1.0 |
| Glyma13g32920 | Defense related | 1,315.2 | 7.4 | 3.3 |
| Glyma04g42870, Glyma06g11890 | PsbS | 1,903.2 | 0.8 | 1.2 |
| Glyma05g00620 | PsaF | 8,936.1 | 0.9 | 1.7 |
G. max locus identifiers—http://www.phytozome.net/cgi-bin/gbrowse/soybean/. Where two gene IDs are listed, cDNA-specific primers amplify both.
MGD, monogalactosyldiacylglycerol synthase; EMB1473, embryo defective 1473; SBPase, sedoheptulose-1,7-bisphosphatase; PsbS, subunit S of photosystem II; PsaF, subunit F of photosystem I.
The cDNA/gDNA mixture was diluted 5-fold and used as template for qRT-PCR with the following components: 5.75 μl H2O, 7.5 μl Power SYBR Green master mix (Applied Biosystems), 0.375 μl forward primer, 0.375 μl reverse primer, and 1 μl template. Assays were performed on an Applied Biosystems 7900 HT instrument, with 40 PCR cycles. Dissociation curves were generated at the end of the PCR to confirm specificity of amplification. For each primer pair and species, we amplified three technical replicates from each of three biological replicates.
Amplification efficiencies were estimated using LinRegPCR (Ramakers et al. 2003) for each individual reaction. Mean efficiency per amplicon was used for relative expression estimates. Expression of each target gene (cDNA-specific amplification) was normalized to genome copy number, as estimated by the geometric mean of amplification from the three gDNA-specific targets. Relative genome-normalized expression values (T2/D3, T2/D4, T2/midparent, and D4/D3) were estimated using the relative expression software tool (REST) (Pfaffl et al. 2002).
We confirmed that T2 retains both D3 and D4 homoeologues for all gene targets (both cDNA and gDNA-specific) by the presence of both D3- and D4-specific single nucleotide polymorphisms (SNPs), as revealed by sequence data from the transcript profiling experiment and/or from Sanger sequencing of cDNA and/or gDNA (supplementary fig. S1 and supplementary table S2, Supplementary Material online). Because T2 has twice as many copies of each target gene as the diploids, relative expression per cell in T2 (T2/D3, T2/D4, and T2/midparent) was obtained by multiplying relative expression per genome by two. For comparisons of D3 and D4, expression per genome is equivalent to expression per cell.
Transcriptome-Normalized Expression Assay
Relative expression per transcriptome was measured by RNA-Seq. Leaflets were pooled from six individuals per species, and RNA was isolated using the Qiagen Plant RNeasy kit with on-column DNase treatment. Sequencing was performed using Solexa/Illumina “sequencing by synthesis” with the following modifications. Poly A+ RNA was annealed to high concentrations of random hexamers, reverse transcribed, and ligated to adapters complementary to sequencing primers. The cDNA was then amplified by 20 cycles of PCR and size fractionated on agarose gels. In total, 200-bp amplicons were excised and sequenced by synthesis with reversible terminator nucleotides with cleavable fluorescence.
To process the data for analysis, files were mirrored to an off-instrument computer using the Illumina platform to perform image analysis, base-calling, quality filtering, and per base confidence scores. Sequences where then aligned using GSNAP (Wu and Nacu 2010) against the 8X genome sequence of soybean (Glycine max; version Glyma1, Soybean Genome Project, DoE Joint Genome Institute), which diverged from the common ancestor of D3, D4, and T2 approximately 5 Ma (Innes et al. 2008). Note that soybean, D3, and D4, all of which are 2n = 40, are fully diploidized descendants of an ancestor that underwent a whole genome duplication approximately 10 Ma (Shoemaker et al. 2006). Roughly half of the genes duplicated by this event are retained in duplicate in the soybean genome (Schmutz et al. 2010). Only reads mapping unambiguously to a single copy in the soybean genome were used in this study.
GSNAP was parameterized to allow spliced alignments of the transcript reads to the genomic reference sequences requiring canonical splice sites and allowing introns of up to 10 kbp; alignments were also allowed to include small indels and mismatches but required that at least 30 of the 36 bp in a read were matched. Alignments above this threshold with the highest number of identities were divided into three classes: uniquely aligned reads, low-copy repetitive alignments matching no more than five locations in the reference, and highly repetitive reads matching >5 locations in the reference. The alignments in the first two classes were further processed using the Alpheus pipeline (Miller et al. 2008) for deriving per-gene read counts and sequence polymorphism calls. The boundaries of each gene were taken as the maximal starting and ending positions from any of the transcripts associated with the gene, and any read alignment partially contained with this span was counted toward the expression of that gene in the given sample. Reads from uniquely aligned sequences were used to estimate expression levels after normalizing read counts to account for overall sampling sizes. Transcript abundance per transcriptome for a given gene was estimated as the number of reads unambiguously mapped to that gene per million unambiguously mapped reads generated by that library (reads per million [RPM]). Because all comparisons involved the relative expression of individual genes across species (as opposed to multiple genes within a species), no adjustment for gene length (e.g., RPKM) (Mortazavi et al. 2008) was necessary.
Calculation of Relative Transcriptome Size
We obtained independent estimates of relative transcriptome size (T2/D3, T2/D4, T2/midparent, and D4/D3) from each of the seven genes assayed by qRT-PCR. The expression per cell (qRT-PCR) estimate obtained for each gene was divided by expression per transcriptome (RNA-Seq) for that gene. The mean of these seven independent estimates (and associated standard error [SE]) was taken as the best overall estimate of relative transcriptome size.
Comparison of cDNA Pools from Genome-Normalized and Transcriptome-Normalized Expression Assays
One of the cDNA-specific primer pairs employed in the qRT-PCR assay amplifies two actin loci (supplementary table S1, Supplementary Material online). In order to confirm that the RNA extracted with the Dr. P kit (used for the qRT-PCR assay) was comparable with the RNA extracted with the Qiagen RNeasy kit (used for RNA-Seq), and quantitatively representative of its corresponding transcriptome, expression of the other six genes assayed by qRT-PCR was also normalized to the combined expression of the actin genes, and relative expression ratios for T2 versus each diploid estimated using REST, as above. RNA-Seq unique RPMs for each of the same six genes were then normalized to the same two actin genes (RPMtarget gene/RPMactin). The actin-normalized expression ratios from qRT-PCR were then compared with the actin-normalized expression ratios from RNA-Seq to determine the correlation of actin-normalized expression estimates between the two platforms (supplementary fig. S2, Supplementary Material online).
Estimation of Relative Homoeologue Expression Levels in T2
We checked each nucleotide position within exons for substitutional differences distinguishing D3 from D4 using consensus sequences from the Illumina reads. Only sites covered by at least two reads in both D3 and D4 were used. For each site that differed between D3 and D4 and to which we had aligned at least five reads from the T2 sample, we determined the proportion of D3-type versus D4-type nucleotides sampled. The homoeologue expression ratio for a gene was calculated by averaging the ratios at each diagnostic site weighted by the number of T2 reads aligned across that site.
Estimation of Genome Sizes and Extent of Endopolyploidy
Young, fully expanded leaflets were collected and stored overnight in the dark on wet paper towels. Leaves were finely chopped in an MgSO4 buffer (Arumuganathan and Earle 1991) and passed through a 30-μm mesh filter (Partec CellTrics) to remove large debris. Propidium iodide (15 μl of a 5 μg/μl solution) and RNase (5 μl of a 5 mg/ml solution) were then added to the filtrate. Samples were run on a Coulter Epics XL-MCL flow cytometer. Measurements of fluorescence intensity were made on 3–4 individuals per species. Data were analyzed using WinMDI.
Absolute genome sizes were estimated by cochopping 12.5 mg of leaf tissue with 12.5 mg of leaf tissue from a plant standard of known genome size. Glycine max (2.5 pg/2C) and Z. mays (5.4 pg/2C) were used as standards for the tetraploid and diploids, respectively (Dolezel et al. 2007).
The extent of endoreduplication was estimated by analyzing 25 mg of leaf tissue without an internal standard. Endoreduplication produces peaks in the fluorescence histogram in multiples of the main (2C) peak. The ratio of endoreduplicated nuclei to total nuclei was quantified by dividing the number of nuclei in the endopolyploid peaks by the combined number of nuclei in the primary and endopolyploid peaks.
Data Deposition
RNA-Seq data submission to NCBI Sequence Read Archive pending.
Results
Expression per Genome
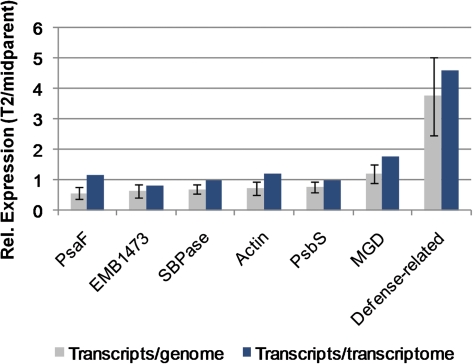
We devised a novel qRT-PCR assay that utilizes gDNA and RNA coextracted from the same tissue to normalize transcript abundance to gDNA abundance. Because RNA and gDNA were extracted from the same cells, in vivo RNA/gDNA ratios were preserved. In addition, we confirmed that amplification efficiencies where comparable (≥1.90) in all three species for each primer pair used in the qRT-PCR assay (data not shown). Consequently, normalizing cDNA amplification by gDNA amplification in qRT-PCR gives a direct readout of transcript abundance per genome. Using this method, we quantified expression per genome in the allotetraploid (T2) and its diploid progenitors (D3 and D4) for seven different genes or gene families (table 1). Across the seven genes/gene families, expression per genome in T2 relative to the midparent value ranged from 0.6× to 3.7× (fig. 2 and supplementary table S3, Supplementary Material online).
FIG. 2.—
qRT-PCR based estimates of transcripts per genome (gray; ± SE; N = 3) and RNA-Seq based estimates of transcripts per transcriptome (blue; N = 1) in T2 relative to the midparent values for seven genes or gene families. Values are ordered by relative expression per genome. The relative number of transcripts per cell in T2 versus midparent is equal to 2× the relative number of transcripts per genome.
Based on RNA-Seq (see below) and/or Sanger sequencing, we confirmed that T2 retains both D3 and D4 homoeologues for each target gene used in the qRT-PCR assay (supplementary fig. S1 and supplementary table S2, Supplementary Material online). Because T2 has two copies of each gene used for genomic normalization for every one copy in the diploids (two homoeologues per diploid gene), we calculated expression per “cell” in T2 relative to its diploid progenitors as two times the relative expression per genome (supplementary table S3, Supplementary Material online).
Expression per Transcriptome
We also profiled the leaf transcriptomes of the allotetraploid (T2) and its diploid progenitor species (D3 and D4) by RNA-Seq. High throughput sequencing using Solexa/Illumina technology generated >5 million 36-bp reads for each species. Reads were uniquely mapped to >35,000 genes in each species, with unique read counts per gene ranging from 1 to >98,000, reflecting the relative abundance of that transcript in the transcriptome (Marioni et al. 2008). The expression level per transcriptome for a given gene was estimated as the number of sequencing reads derived from that gene divided by the total number of reads derived from that sample, reported as RPM. Because we compared the relative expression of individual genes across species (as opposed to multiple genes within a species), relative expression estimates were not affected by variation in gene length, making length adjustments (e.g., RPKM) (Mortazavi et al. 2008) unnecessary. Across the seven genes/gene families for which relative expression per “genome” was determined by qRT-PCR, expression per transcriptome in T2 relative to the midparent value ranged from 0.8× to 4.6× (fig. 2 and supplementary table S3, Supplementary Material online).
Comparison of cDNA Pools from Genome-Normalized and Transcriptome-Normalized Expression Assays
Because the cDNA template used in the qRT-PCR assay was generated in a nonstandard way (reverse transcription was performed on RNA in a native mixture with gDNA), we verified that these cDNA pools were quantitatively equivalent to the cDNA pools used for RNA-Seq. Following standard qRT-PCR methodology, expression estimates obtained using the TNA-derived cDNA for 6 of the 7 genes examined were normalized to the expression of actin (the seventh gene family), and relative expression ratios for T2 versus the diploid midparent value were estimated. RNA-Seq RPMs for each of the same six genes were then normalized to the same actin genes (RPMtarget gene/RPMactin). The actin-normalized expression ratios from qRT-PCR were then compared with the actin-normalized expression ratios from RNA-Seq. Across the six genes, a strong correlation was observed between the two estimates (Pearson correlation coefficient = 0.99; supplementary fig. S2, Supplementary Material online), indicating that the RNA-Seq and qRT-PCR cDNA preps were equivalently representative of the transcriptomes from which they were derived.
Relative Transcriptome Size
To estimate the size of the tetraploid transcriptome relative to each diploid transcriptome, we then divided the per cell expression ratios from the quantitative polymerase chain reaction (qPCR) assay by the per transcriptome expression ratios from the RNA-Seq data set (fig. 1). The logic of this calculation can be seen algebraically. The qPCR result gives the expression of a gene in the tetraploid relative to the expression in the diploid on a per cell basis (fig. 1):
The RNA-Seq result gives the expression in the tetraploid relative to the expression in the diploid on a per transcriptome basis (fig. 1):
 |
Dividing ratio 1 by ratio 2 yields the following:
This is the size of the tetraploid transcriptome relative to the size of the diploid transcriptome.
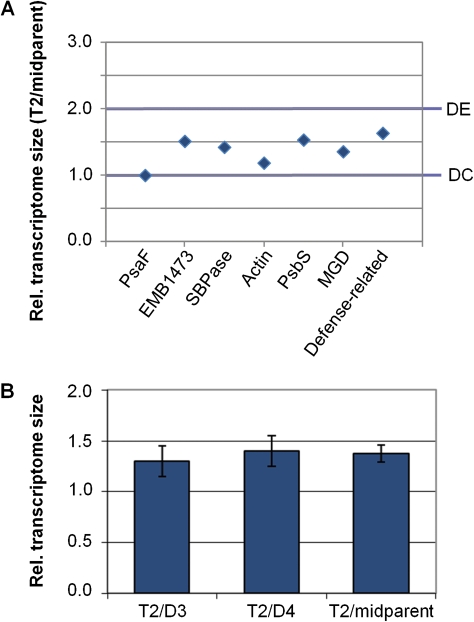
With this approach, we obtained seven independent estimates of the size of the tetraploid transcriptome relative to each diploid progenitor transcriptome and to the diploid midparent transcriptome (fig. 3A; supplementary table S3, Supplementary Material online). There was variation among individual gene estimates, as might be expected given that there is error associated with both RNA-Seq and qPCR data, but all estimates for T2/midparent fell between 1- and 2-fold (the expected values if the T2 transcriptome overall was dosage compensated or exhibited 1:1 dosage effects, respectively). With these data, we rejected the null hypothesis that the T2 transcriptome was doubled (a genome-wide dosage effect) relative to the midparent transcriptome (P = 0.0002; One-sample t-test), as well as the null hypothesis that the T2 transcriptome was equal in size (genome-wide dosage compensation) to the midparent transcriptome (P = 0.0031; One-sample t-test). On a global scale, therefore, the T2 leaf transcriptome has been partially dosage compensated. Our data indicated that the leaf transcriptome of the tetraploid under these conditions was 1.4-fold (±0.1 SE) larger than the midparent transcriptome (fig. 3B) and 1.3- to 1.4-fold (±0.2 SE) larger than the transcriptomes of either individual diploid progenitor. The diploid transcriptomes did not differ significantly in size (P = 0.7561; one-sample t-test). We estimated that the D4 leaf transcriptome was 1.1-fold (±0.2 SE) larger than the D3 leaf transcriptome (fig. 3B).
FIG. 3.—
T2 transcriptome size relative to the transcriptomes of its diploid progenitors. (A) Seven individual gene-based estimates of relative transcriptome size (T2 vs. the diploid midparent transcriptome).“DE” designates the expected value if the T2 transcriptome experienced genome-wide 1:1 dosage effects. “DC” designates the expected value if the T2 transcriptome experienced genome-wide dosage compensation. (B) Average estimate of tetraploid transcriptome size relative to the transcriptomes of each diploid progenitor and to the midparent diploid transcriptome (±SE; N = 7).
Endopolyploidy (the occurrence of different ploidy levels within different cells of an organism) is common in seed plants (Barow 2006). Because our transcriptome size estimates were obtained by normalizing gene expression to ploidy level (genome copy number), differences in the extent of endopolyploidy between T2 and D3 or D4 would affect our estimates of transcriptome size. In order to quantify the extent of endopolyploidy in D3, D4, and T2, we performed flow cytometry on leaf tissue of a comparable developmental stage (young, fully expanded) as was used for RNA-Seq and qRT-PCR. We observed minimal levels of endopolyploidy in all three species, with comparable fractions of endopolyploid nuclei in each (4–7% of nuclei; supplementary table S4 and supplementary fig. S3, Supplementary Material online). Our estimates of transcriptome size are not, therefore, skewed by differences in endopolyploidy.
Dosage Responses Across the Tetraploid Transcriptome
Once an estimate of transcriptome size was obtained, estimates of dosage response could then be made for each gene in the transcriptome profiling data set. Because the T2 transcriptome was estimated to be 1.4-fold (±0.1 SE) larger than the midparent diploid transcriptome, a gene that has undergone complete dosage compensation in T2 would exhibit a transcriptome-normalized expression level of 0.7 times the midparent diploid level (0.7× diploid copies per transcriptome × 1.4 diploid transcriptome equivalents per cell ≈1.0× diploid copies per cell or 0.5× copies per genome). Likewise, a gene whose expression has experienced a 1:1 dosage effect would exhibit a transcriptome-normalized expression level of 1.4× the midparent level (1.4 × 1.4 ≈ 2.0× copies per cell or 1.0× copies per genome). Based on the SE associated with our estimate of transcriptome size (±0.1), a 95% confidence interval (CI) for the size of the T2 transcriptome relative to the midparent value is approximately 1.2- to 1.6-fold (1.4 ± 1.96 × SE). From this, we approximated CIs for each response: genes exhibiting transcriptome-normalized expression in T2 between 0.6 and 0.8× the midparent level were most likely dosage compensated (0.6 × 1.6 ≈ 1.0; 0.8 × 1.2 ≈ 1.0) and genes exhibiting transcriptome-normalized expression between 1.3 and 1.7× the midparent level most likely exhibited a 1:1 dosage effect (1.3 × 1.6 ≈ 2.0; 1.7 × 1.2 ≈ 2.0).
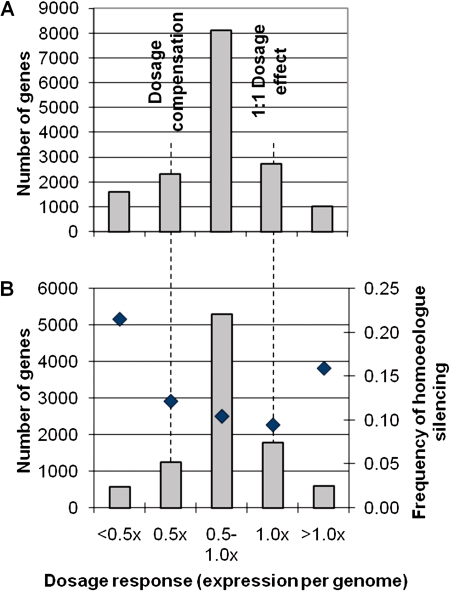
Figure 4A shows the distribution of dosage responses in T2. Of 15,761 genes in our RNA-Seq data set with at least 10 uniquely mapped RPM in at least 1 of the 3 species, 2,319 (14.7%) exhibited transcriptome-normalized expression in T2 consistent with dosage compensation, and 2,724 (17.3%) exhibited expression levels consistent with a 1:1 dosage effect. The majority of genes in T2 (8,115; 51.5%) displayed an intermediate dosage response (0.8–1.3× midparent). Of the remaining genes, 1,583 genes (10.0%) exhibited a negative dosage effect (<0.5× diploid expression per genome), and 1,020 genes (6.5%) exhibited a greater than 1:1 dosage effect (>1× diploid expression per genome).
FIG. 4.—
Genome-wide distribution of gene dosage responses and fraction of genes within each dosage response category exhibiting homoeologue silencing in the T2 allotetraploid. (A) Number of genes from the RNA-Seq data set with ≥10 unique RPM in at least 1 of the 3 species showing specified dosage responses in T2. (B) Of the genes from panel A for which homoeologue expression could be estimated, number of genes showing specified dosage responses (gray bars) and the fractions of each for which one homoeologue is silenced under these conditions (♦). Dosage responses are expressed as relative expression per genome (T2/midparent): 0.5× = dosage compensation, 1.0× = 1:1 dosage effect.
Homoeologue Modulation
RNA-Seq enables estimates of the contributions of each homoeologue to total expression in the T2 tetraploid (see supplementary fig. S1, Supplementary Material online). Combining this information with our estimates of dosage response, we could identify patterns of homoeologue deployment associated with each dosage response category. For example, dosage compensation could be achieved by silencing 1 of 2 homoeologues while maintaining the other at its diploid expression level or by downregulating both copies. Of the 2,319 genes that were considered to be dosage compensated, homoeologue contributions could be determined for 1,240. Of these 1,240 genes, 151 (12.2%) expressed only one homoeologue (fig. 4B). By comparison, 168/1,772 (9.5%) genes that exhibited a 1:1 dosage effect expressed only 1 of 2 homoeologues (fig. 4B). Thus, there was a slight but significant increase in the frequency of homoeologue silencing among dosage-compensated genes versus genes that showed a 1:1 dosage effect (χ12 = 5.60, P = 0.02). Nonetheless, even among dosage-compensated genes, the vast majority expressed both homoeologues, indicating that in most cases dosage compensation was achieved by more subtle modulations of homoeologue expression.
As might be expected, genes that exhibited negative dosage effects (<0.5× diploid expression per genome) silenced homoeologues at the highest frequency (21.5% of 572 genes; fig. 4B), which was significantly higher than for genes that were dosage compensated (χ12 = 26.53, P < 0.0001). Surprisingly, the next highest category of genes with one silenced homoeologue was the group of genes showing >1:1 dosage effects (15.9% of 603 genes), which was also significantly higher than the group of genes that were dosage compensated (χ12 = 4.90, P = 0.03). Thus, many loci showing strongly upregulated expression in T2 versus its diploid progenitors did so using only 1 of 2 homoeologues. Overall, a pattern emerged in which genes showing the most extreme dosage responses in either direction (<0.5× or >1.0× diploid expression per genome) were more likely to exhibit homoeologue silencing than genes showing intermediate responses (0.5–1.0× diploid expression per genome; χ12 = 66.73, P < 0.0001; fig. 4B).
Transcriptome Versus Genome Size
Using flow cytometry, we estimated the T2 genome to be 1.89-fold larger than the midparent genome (1.84-fold larger than D3 and 1.93-fold larger than D4) or 94.5% of the sum of the two progenitor genomes (supplementary table S4, Supplementary Material online). Of 10,311 genes with sufficient depth of sequence coverage in the RNA-Seq data set and diagnostic SNPs distinguishing D3 and D4 (supplementary fig. S1, Supplementary Material online) to estimate homoeologue expression, 8,934 (86.8%) had sequences derived from both homoeologues in T2. Thus, homoeologues were retained for at least ∼87% of genes initially duplicated in T2 (and almost certainly more because some homoeologues are likely retained but not expressed highly enough under these conditions to be detected). Consequently, we estimated that T2 has 1.87–2.0 homoeologues per diploid gene (i.e., 1.87–2.0 times the number of genes per cell) but only 1.4 times the number of transcripts per cell (fig. 3B). Averaged across the genome, therefore, expression per gene in T2 is approximately 0.70- (1.4/2.0) to 0.75-fold (1.4/1.87) that of its diploid progenitors.
Discussion
Because transcript profiling experiments yield transcriptome-normalized expression values, they provide no information about expression per cell without knowing the relative sizes of the transcriptomes being compared. Here, we have described a novel qRT-PCR assay that provides direct estimates of expression per genome and per cell and have shown how these estimates can be coupled with transcript profiling data to obtain estimates of relative transcriptome size. These estimates can in turn be used to determine relative expression per cell for every gene in the transcript profiling data set.
Kanno et al. (2006), recognizing the same problem, proposed an alternative method to determine expression level per cell but did not utilize their data to estimate relative transcriptome sizes. Also, because their focus was on normalizing microarray data, their method is necessarily less direct than ours (they used spiked RNA as a proxy for the gDNA initially present in the sample as opposed to the gDNA itself) and would require precise quantification of genome sizes before being applied to cross-species or cross-ploidy level comparisons. In contrast, the method described here is insensitive to genome size and only requires knowledge of target gene and genome copy number per cell (ploidy level).
Allopolyploidy and Transcriptome Size
By coupling transcript profiling data with a genome-normalized qRT-PCR assay, we have provided the first estimates of transcriptome size (number of transcripts per cell) for several closely related species: a tetraploid and its diploid progenitors. Whereas the two diploid leaf transcriptomes are approximately the same size, that of the tetraploid is significantly larger. But despite the fact that the T2 tetraploid (G. dolichocarpa) is of fairly recent origin (within the last 100,000 years) and retains ≥87% of its genes in duplicate, its leaf transcriptome is only ∼1.4-fold larger than the transcriptomes of its diploid progenitors.
It is possible that the T2 leaf transcriptome was doubled initially and has subsequently undergone downsizing in a process akin to genome diploidization. If so, because we observe an approximately 30% reduction in transcriptome size (vs. the sum of the two diploid transcriptomes), but only a 6% reduction in genome size (vs. the sum of the two diploid genomes), and ≤7% reduction in gene copy number, this suggests that transcriptome downsizing has progressed to a greater degree than genome downsizing in this species. The transcriptome may have experienced immediate and widespread dosage compensation upon genome doubling, perhaps via epigenetic mechanisms—changes in DNA methylation have been observed in other polyploid species in the first generations following doubling (Lee and Chen 2001; Kashkush et al. 2002; Madlung et al. 2005), and chromatin modifications (histone acetylation and methylation) are associated with changes in expression of FLC (Wang et al. 2006b), CCA1, and LHY (Ni et al. 2009) in synthetic Arabidopsis allotetraploids. Estimating transcriptome sizes in natural polyploids of various ages, as well as in synthetic polyploids, will shed light on this question and reveal if changes in cellular transcript abundance are consistent across species or if they are lineage specific. Additionally, because transcriptomes vary by tissue type and growth condition, it remains to be determined whether other tissues or conditions exhibit similar responses in terms of transcriptome size.
Dosage Responses of Individual Genes
To date, dosage responses associated with polyploidy have only been estimated for 18 genes in a synthetic maize autopolyploid series (Guo et al. 1996). With an estimate of relative transcriptome size in hand, we were able to infer dosage responses for 15,761 genes in T2 (fig. 3A). In contrast to the overall pattern observed in maize (Guo et al. 1996), in which the majority of genes surveyed exhibited a 1:1 dosage effect, the majority of genes in the T2 allopolyploid (8,115; 51.5%) display an intermediate dosage response (0.8–1.3× midparent), driving the genome-wide average of partial dosage compensation. Only about 17% of the genes in T2 exhibit a 1:1 dosage response.
This difference in global dosage response pattern could be due to the hybrid origin of T2. Whereas dosage responses in maize were examined in an autopolyploid series (Guo et al. 1996), T2 was formed via interspecific hybridization, producing novel combinations of cis- and trans-acting transcriptional regulators. Alternatively, some of the observed differences may be due to gene expression evolution in T2. Despite a relatively recent origin, T2 has been subject to natural selection for tens of thousands of years, whereas the maize polyploids were studied in the first generations following synthesis in the laboratory.
It is also possible that the limited sampling in maize (18 genes) does not provide a representative picture of overall dosage responses. Application of the methods described here to the maize synthetic autopolyploid system, as well as to other polyploidy model systems, would give a more comprehensive picture of the similarities and differences in dosage response patterns between natural and synthetic polyploids as well as between auto- and allopolyploids.
Modulation of Homoeologue Expression Across an Allopolyploid Genome
The contributions of D3 and D4 homoeologues to T2 expression could be determined for genes in which D3- or D4-specific SNPs were sequenced (supplementary fig. S1, Supplementary Material online). Thus, we were able to explore patterns of homoeologue deployment under each dosage response. In most cases, both homoeologues were expressed, even when total expression was modulated to the midparent diploid level or less. Overall, 1 of 2 copies was silent for 11.5% of the homoeologue pairs examined. In a study of homoeologue expression biases in ovules of a natural cotton allotetraploid (Adams et al. 2003), only 1 of 40 pairs (2.5%) exhibited complete silencing. A more recent study using a homoeologue-specific microarray to survey the same cotton allotetraploid more broadly (Flagel et al. 2008) observed homoeologue silencing for 115 of 1,383 genes (8.3%). Thus, absolute silencing of homoeologues may be relatively rare.
Though generally uncommon, our data indicate that the frequency of homoeologue silencing varies significantly by dosage response (fig. 3B). The group of genes exhibiting dosage compensation (expression per cell equal to the midparent diploid expression level) had a higher frequency of homoeologue silencing than genes exhibiting a 1:1 dosage effect (expression per cell double that of the midparent diploid expression level). Additionally, genes exhibiting extreme dosage responses in either direction (<0.5× per genome or >1.0× per genome) were significantly more likely to silence one homoeologue (21.5% and 15.9%, respectively) than genes that have undergone more moderate dosage responses (0.5× to 1.0× per genome). For genes that have experienced a negative dosage effect (expression below the diploid level per cell), this makes intuitive sense. For genes that have experienced a >1:1 dosage effect, however, this result is surprising. In these cases, the polyploid is producing more than double the midparent number of transcripts per cell from the same number of loci as its diploid progenitors. Thus, complete silencing of one homoeologue is accompanied by strong upregulation of the other.
Relevance and Utility of Overall Transcriptome Size
Normalizing expression data per cell provides a reliable means to compare transcript profiling experiments performed with different RNA samples and on different platforms (Kanno et al. 2006). In addition, quantifying relative expression per cell is necessary to understand gene dosage responses and has the potential to reveal biologically significant differences in gene regulation that may be obscured in transcriptome-normalized data.
Equivalent analyses of transcriptome size would give greater context to existing (Hegarty et al. 2005, 2006, 2008; Wang et al. 2006a) and future transcript profiling experiments comparing species and ploidy levels by making it possible to determine if additivity on a per transcriptome basis (i.e., equal transcriptome-normalized expression) translates to additivity in absolute expression. At present, different studies of gene expression in polyploids operate on the assumption that “additive” transcriptome-normalized expression represents either midparent expression (i.e., dosage compensation) or the sum of expression from the two diploids—that is, a 1:1 dosage effect, and often the two are used interchangeably (Jackson and Chen 2009), despite very different meanings. As our data show, either assumption could be faulty.
Recent genomic studies have led to renewed interest in gene dosage evolution (Papp et al. 2003; Blanc and Wolfe 2004; Freeling and Thomas 2006; Paterson et al. 2006; Thomas et al. 2006). Reciprocal patterns of duplicate retention following polyploidy and nonpolyploid duplications suggest that dosage sensitivity is, in many cases, driving gene family evolution (Freeling 2009; Birchler and Veitia 2010). Dosage sensitivity correlates with the extent to which a gene’s product forms protein–protein interactions, and the balance hypothesis correctly predicts that such “connected” genes (Thomas et al. 2006) will tend to retain polyploidy duplicates and eliminate nonpolyploid duplicates. There are, however, numerous exceptions. Genes that appear to meet the criteria of being connected but do not follow the predictions of the balance hypothesis may represent genes for which transcript abundance is readily decoupled from gene dosage (Veitia et al. 2008; Edger and Pires 2009). Consequently, cataloging dosage responses across the genome, as we have done here, will help to test and refine the balance hypothesis.
Finally, the qPCR approach utilized here, using gDNA to normalize expression estimates, could provide more reliable results than the typical alternative of normalizing to expression of a single reference gene in any instance where relative expression estimates are needed. Nicot et al. (2005) evaluated the stability of expression of seven housekeeping genes commonly used for RT-PCR normalization and found significant variation in expression in response to various stresses. They concluded that only 1 of the 7 (“Elongation factor 1-α”; Elf1α) was suitable as an internal reference for the three stresses they examined. Even Elf1α, however, showed a 2–3 cycle range in threshold cycle (Ct) between control and cold stress conditions. Variation in the expression level of housekeeping genes has led some to recommend using combinations of genes as internal controls (Thellin et al. 1999; Vandesompele et al. 2002). This approach, however, greatly increases the size and complexity of an RT-PCR experiment.
In contrast, normalizing to gene copy number may be simpler and more reliable. Gene copy number is more stable than gene expression and, consequently, provides a better reference for normalization. This would be true for all types of comparisons but particularly in the case of cross-species or cross-ploidy level comparisons, where the expression levels of individual housekeeping genes might differ considerably. In a recent study of the effects of ploidy and hybridization on the circadian clock, expression estimates of central oscillator genes were normalized using Actin2 (ACT2) expression (Ni et al. 2009). The possibility for variation in ACT2 expression arising from genome doubling or hybridity was not discussed but is potentially significant. In the present study, RNA-Seq data indicate that the combined expression of two ACT2 orthologs in the T2 tetraploid is 1.4× the D4 diploid level on a per transcriptome basis. Thus, normalizing to Actin would tend to exaggerate apparent cases of downregulation and obscure genuine cases of upregulation associated with polyploidy in T2. Genomic copy number is more stable than expression level (though differences in endoreduplication must be accounted for) and, arguably, more easily verified. Consequently, gene copy normalization should provide more reliable estimates of relative expression, with the added advantage of providing direct information about dosage responses.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank Gregory D. May and Andrew D. Farmer of the National Center for Genome Resources for carrying out Illumina sequencing and primary data processing and Daniel C. Ilut of Cornell University for writing scripts to calculate homoeologue expression levels using the RNA-Seq data. We are grateful to Brandon Gaut and three anonymous reviewers for comments on the manuscript. This work was supported by The National Science Foundation (grant numbers IOS-0744306 and DEB-0709965 to J.J.D.).
References
- Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci U S A. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MR, et al. A trispecies aspergillus microarray: comparative transcriptomics of three aspergillus species. Proc Natl Acad Sci U S A. 2008;105:4387–4392. doi: 10.1073/pnas.0709964105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep. 1991;9:217–229. [Google Scholar]
- Barow M. Endopolyploidy in seed plants. Bioessays. 2006;28:271–281. doi: 10.1002/bies.20371. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 2010;186:54–62. doi: 10.1111/j.1469-8137.2009.03087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LY, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Rauscher JT, Brown AHD. Evolution of the perennial soybean polyploid complex (glycine subgenus glycine): a study of contrasts. Biol J Linn Soc Lond. 2004;82:583–597. [Google Scholar]
- Edger P, Pires J. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17:699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- Ehrendorfer F. Polyploidy and distribution. In: Lewis WH, editor. Polyploidy: biological relevance. New York: Plenum; 1980. pp. 45–60. [Google Scholar]
- Enard W, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc Natl Acad Sci U S A. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L, Udall J, Nettleton D, Wendel J. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 2008;6:16. doi: 10.1186/1741-7007-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Borevitz J. Using DNA microarrays to study natural variation. Curr Opin Genet Dev. 2006;16:553–558. doi: 10.1016/j.gde.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Pritchard JK, Thornton K. Characterizing natural variation using next-generation sequencing technologies. Trends Genet. 2009;25:463–471. doi: 10.1016/j.tig.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. Plant speciation. New York: Columbia University Press; 1981. [Google Scholar]
- Guo M, Davis D, Birchler JA. Dosage effects on gene expression in a maize ploidy series. Genetics. 1996;142:1349–1355. doi: 10.1093/genetics/142.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JP, et al. A comparison of the Thlaspi caerulescens and Thlaspi arvense shoot transcriptomes. New Phytol. 2006;170:239–260. doi: 10.1111/j.1469-8137.2006.01662.x. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Hiscock SJ. Genomic clues to the evolutionary success of polyploid plants. Curr Biol. 2008;18:R435–R444. doi: 10.1016/j.cub.2008.03.043. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, et al. Changes to gene expression associated with hybrid speciation in plants: further insights from transcriptomic studies in senecio. Philos Trans R Soc Lond B Biol Sci. 2008;363:3055–3069. doi: 10.1098/rstb.2008.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty MJ, et al. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, et al. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Mol Ecol. 2005;14:2493–2510. doi: 10.1111/j.1365-294x.2005.02608.x. [DOI] [PubMed] [Google Scholar]
- Hovav R, Chaudhary B, Udall JA, Flagel L, Wendel JF. Parallel domestication, convergent evolution and duplicated gene recruitment in allopolyploid cotton. Genetics. 2008a;179:1725–1733. doi: 10.1534/genetics.108.089656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R, et al. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc Natl Acad Sci U S A. 2008b;105:6191–6195. doi: 10.1073/pnas.0711569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RW, et al. Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 2008;148:1740–1759. doi: 10.1104/pp.108.127902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Chen ZJ Forthcoming. Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol. 2009;13:153–159. doi: 10.1016/j.pbi.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno J, et al. “Per cell” normalization method for mRNA measurement by quantitative PCR and microarrays. BMC Genomics. 2006;7:64. doi: 10.1186/1471-2164-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chen ZJ. Protein-coding genes are epigenetically regulated in arabidopsis polyploids. Proc Natl Acad Sci U S A. 2001;98:6753–6758. doi: 10.1073/pnas.121064698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis WH. Polyploidy in species populations. In: Lewis WH, editor. Polyploidy: biological relevance. New York: Plenum; 1980. pp. 103–144. [Google Scholar]
- Madlung A, et al. Genomic changes in synthetic arabidopsis polyploids. Plant J. 2005;41:221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NA, et al. Management of high-throughput DNA sequencing projects: Alpheus. J Comput Sci Syst Biol. 2008;1:132–148. doi: 10.4172/jcsb.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Paterson AH, et al. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 2006;22:597–602. doi: 10.1016/j.tig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeil BE, Craven LA, Brown AHD, Murray BG, Doyle JJ. Three new species of northern Australian glycine (Fabaceae, Phaseoleae), G. gracei, G. montis-douglas and G. syndetika. Funct Plant Biol. 2006;19:245–258. [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rapp R, Udall J, Wendel J. Genomic expression dominance in allopolyploids. BMC Biol. 2009;7:18. doi: 10.1186/1741-7007-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Rokas A, Abbot P. Harnessing genomics for evolutionary insights. Trends Ecol Evol. 2009;24:192–200. doi: 10.1016/j.tree.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter J, Doyle JJ. Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol. 2006;9:104–109. doi: 10.1016/j.pbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold; 1971. [Google Scholar]
- Tang H, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellin O, et al. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall JA, Swanson JM, Nettleton D, Percifield RJ, Wendel JF. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics. 2006;173:1823–1827. doi: 10.1534/genetics.106.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034.1–research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Lee H, Chen ZJ. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in arabidopsis allopolyploids. Genetics. 2006b;173:965–974. doi: 10.1534/genetics.106.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Genomewide nonadditive gene regulation in arabidopsis allotetraploids. Genetics. 2006a;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, et al. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S Forthcoming. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]