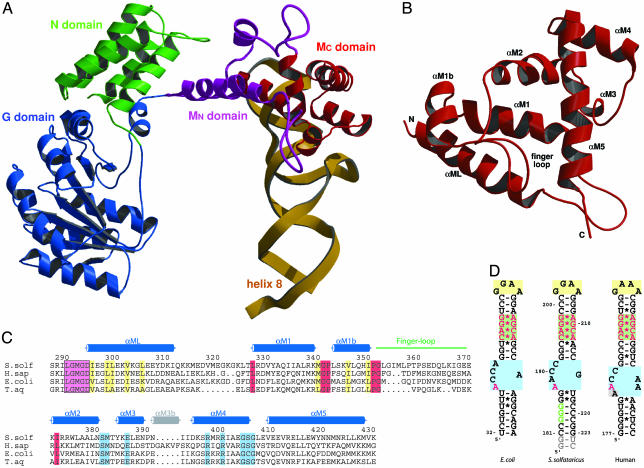
Fig. 1.
Overall structure of the SRP core. (A) Domain arrangement in SRP54 in complex with SRP RNA helix 8 shown in a ribbon representation of the N (green), G (blue), and M (purple/red) domains and the SRP RNA helix 8 (yellow). The novel N-terminal part of the M domain (MN) containing the linker helix (αML) and the closed finger loop is highlighted in purple. (B) Ribbon diagram of the M domain in a top view compared with A. The finger loop on top is folded into the hydrophobic groove, which is lined by helices αM1, αM1b, αM2, and αM5. (C) Structure-based sequence alignment of the M domain from different species (S. solf, S. solfataricus; H. sap, Homo sapiens; T. aq, T. aquaticus). Residue numbering corresponds to S. solfataricus SRP54. The secondary structure elements are indicated above the alignment. The highly conserved LGMGD motif (in a magenta box) and the long amphipathic linker helix (αML) are indicated. Regions involved in adjusting the finger loop for signal peptide binding are marked in red. Highly conserved hydrophobic residues involved in the contact with the N domain are shown in yellow. Residues involved in RNA binding are shown in blue. The short helix αM3b (gray) is found only in higher eukaryotes. (D) Nucleotide sequences of helix 8 of the SRP RNA from S. solfataricus used for this work (Center) compared with E. coli (Left) domain IV and human (Right) helix 8. The conserved features of helix 8 are highlighted: GNRA-type tetraloop (yellow box), symmetric loop (green box), and asymmetric loop (blue box). Nucleotides equivalent to the ones protected by the NG domain in A. fulgidus (179-GGG in A. fulgidus) (52) are in green letters, and nucleotides involved in SRP54M binding are highlighted in red. Non-native nucleotides added from the T7 promotor and for ribozyme cleavage are marked in gray.