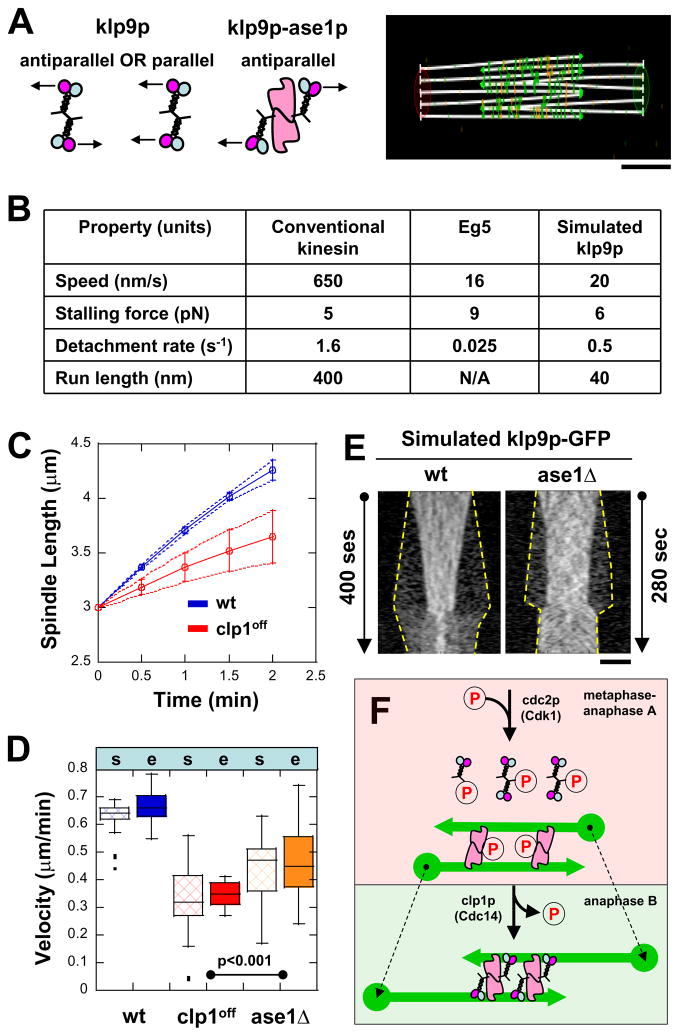
Figure 7. Cytosim model of how klp9p (motor) and ase1p (MAP) interact for proper anaphase B.
(A) Cytosim model assumption: klp9p is either parallel or antiparallel by itself, but is antiparallel when bound to ase1p. Bar, 1 μm.
(B) Parameters used in simulations (see text). Parameters of klp9p are assumed to be similar to Eg5, a similar mitotic kinesin.
(C) Comparative plot of spindle length vs. time and box plot of anaphase B spindle elongation velocity of simulated wildtype cells (wt, blue) and mutant clp1off cells (clp1off, red). Simulation reproduces the experimental results (compare with Fig. 4C).
(D) Comparative box plot of simulated (s) and experimental (e) spindle velocities for wildtype cells (wt, blue), clp1off cells (clp1off, red), and ase1Δ cells (ase1Δ, orange). Spindle velocities of ase1Δ cells are intermediate between wt and clp1off.
(E) Comparative kymographs of simulated klp9p-GFP focusing at the middle of the spindle in wildtype and ase1Δ cells. Dashed yellow lines show positions of the spindle poles. In the absence of ase1p, klp9p is less focused at the spindle midzone, similar to experimental findings (compare with Fig. 3B and 5C). Bar, 1 μm.
(F) Model of how kinase cdc2p (Cdk1) and phosphatase clp1p (Cdc14) regulate the function of klp9p (motor) and ase1p (MAP) for proper anaphase B.