Abstract
The synergistic activity between nitric oxide (NO) released from diazeniumdiolate-modified proline (PROLI/NO) and silver (I) sulfadiazine (AgSD) was evaluated against Escherichia coli, Enterococcus faecalis, Proteus mirabilis, Pseudomonas aeruginosa, Staphylococcus aureus and Staphylococcus epidermidis using a modified broth microdilution technique and a checkerboard-type assay. The combination of NO and AgSD was defined as synergistic when the fractional bactericidal concentration (FBC) was calculated to be <0.5 Gram-negative species were generally more susceptible to the individual antimicrobial agents than the Gram-positive bacteria. The in vitro synergistic activity of AgSD and NO observed against a range of pathogens strongly supports future investigation of this therapeutic combination, particularly for its potential use in the treatment of chronic and burn wounds.
Keywords: antimicrobial, combination therapy, nitric oxide, silver sulfadiazine, synergy, topical
Introduction
The antimicrobial properties of silver and more specifically ionic silver (Ag+) have been recognized and utilized for centuries. Nano- to micromolar concentrations of Ag+ exhibit broad-spectrum bactericidal, fungicidal, vircidal, and protozoicidal activity,1 bonding covalently to electron-donating groups (e.g., the sulfhydryl of cysteine) or electrostatically to negatively-charged molecules (e.g., DNA). Most bacterial sites targeted by Ag+ are proteinaceous, where alterations in amino acid residues lead to structural damage and disruption of replicative and metabolic processes, ultimately resulting in cell death.1–6 Evidence suggests that interactions with DNA also play an important role in the antimicrobial efficacy of Ag+.4, 5 Generally applied as an external treatment, Ag+ is delivered via silver compounds such as silver (I) sulfadiazine (AgSD).7 Most causative pathogens related to burns and chronic wound infections are susceptible to the levels of AgSD attainable topically.8, 9 Fortunately, exposure to clinical levels of Ag+ typically does not pose a threat to human health, despite the broad range of reactivity.1, 7
Like Ag+, nitric oxide (NO) is a broad-spectrum antimicrobial agent that has a number of cellular targets and the endogenous expression of NO has been conserved throughout higher organisms as the immune system’s first-line defense against infection.10–13. Although NO can modify proteins and other biological macromolecules directly,14 it is a highly reactive radical and frequently combines with locally abundant small molecules such as oxygen (O2) and superoxide (O2−), generating an arsenal of reactive byproducts that include dinitrogen trioxide (N2O3) and peroxynitrite (ONOO−).10, 15 Collectively these reactive species evoke potent antibacterial effects by rendering nitrosative and oxidative stresses to bacteria.10, 15–19 In vitro, NO administered via soluble, small-molecule diazeniumdiolate NO donors, from nanoparticle delivery vehicles,20–22 and from NO-releasing xerogel coatings.23–25 has been shown to kill a range of pathogens.
Although the appropriate use of antimicrobials to treat infection is a beneficial practice, artificial pressures resulting from over use, patient non-compliance, and widespread application have promoted the unnatural selection of inherently resistant microbes.26 Even resistance to broad-spectrum agents has been observed.7, 27 To complicate matters, methods of gene sharing employed by bacteria promote the localized collection of resistance determinants, often on transferable plasmids, leading to the emergence of multi-drug resistant (MDR) and extensively drug resistant (XDR) species.28, 29 In recent years, a dramatic rise in the incidence of ‘super bugs,’ or bacteria that are resistant to those antibiotics generally reserved as a last resort treatment option (e.g. vancomycin), has been observed.30, 31 Unfortunately, the discovery of new antimicrobials progresses slowly while resistance factors to all clinically employed antimicrobial agents emerge rapidly,26 creating a critical need for alternative approaches to treating infection.
Combination therapy is one strategy for stemming the emergence of resistant species.32–34 The concerted use of two or more biocides with different mechanisms of action decreases the likelihood that an organism will possess all the traits necessary to ensure its selection and survival. Agents possessing a broad spectrum of antimicrobial action may both lower the probability of developing resistance and manage the polymicrobial burden typically found in topical infections.33, 34 As smaller quantities of each drug are generally required in the application of combination therapy, dose-related toxicity experienced to a particular biocide may also be reduced.33, 34 In the best case scenario, the combination of antimicrobials results in synergistic activity.33, 34 Synergy occurs when two agents working in concert exert a greater than additive effect, resulting in combinations that are more potent than equivalent doses administered individually, further reducing potential toxicity to the patient and cost of treatment. Herein, NO generated from diazeniumdiolate-modified proline (PROLI/NO) and AgSD were evaluated alone and in combination against 9 pathogenic microbes using acute (2 h) time-kill viability assays. The synergistic in vitro effect of both agents was assessed for four Gram-negative and five Gram-positive strains of bacteria, including two antibiotic-resistant ‘super bugs.’
Materials and methods
Materials
L-proline, sulfadiazine (SD), and AgSD were obtained from Sigma-Aldrich (St. Louis, MO). Tryptic soy broth (TSB) and tryptic soy agar (TSA) were manufactured by BD (Franklin Lakes, NJ) and purchased from Fisher Scientific (Pittsburgh, PA). Sodium chloride, potassium chloride, and sodium phosphate monobasic obtained from Fisher and sodium phosphate dibasic obtained from Sigma-Aldrich were used to prepare phosphate buffered saline (PBS, Ic = 0.16 M, pH = 7.4). Distilled water was purified using the Millipore Milli-Q UV Gradient A-10 system (Bedford, MA) to a resistivity of 18.2 MΩ cm and used to prepare the reagents for bactericidal assays. Materials used for growing pathogens and/or evaluating antimicrobial activity were exposed to UV radiation or sanitized in an autoclave prior to use, unless purchased sterile. Argon, NO, nitrogen (N2), and a NO standard (25.7 ppm in N2) were purchased from National Welders (Raleigh, NC).
Synthesis and characterization of PROLI/NO
The synthesis protocol reported by Saavedra, et al. was used in the preparation of PROLI/NO.35 Briefly, 10 g of L-proline was dissolved in 39 mL of 25% sodium methoxide in methanol. An additional 20 mL of methanol was added, and the solution was placed into a custom NO reaction bomb, which was then purged with Ar. The proline solution was then exposed to 5 atm of NO for 3 d to form PROLI/NO as a white precipitate in the methanol. After purging the bomb with Ar, the precipitate was isolated by vacuum filtration, washed with ether, and dried under vacuum. The white solid (PROLI/NO) was divided into small aliquots (< 1 g) and stored over dessicant at −20 °C.
Nitric oxide release from PROLI/NO was characterized using a chemiluminescent NO analyzer (Sievers Model 280, Boulder, CO). Briefly, a known quantity of PROLI/NO was inserted into a glass flask containing PBS at 37 °C. Nitric oxide generated into solution via diazeniumdiolate NO donor decomposition was carried to the analyzer by N2 bubbling through the solution at a flow rate of 80 mL min−1. The NO analyzer was calibrated using an atmospheric sample passed through an NO zero filter and a 25.7 ppm NO standard. The NO release from PROLI/NO was measured periodically to ensure no significant decomposition of the NO donor over the duration of use.
Bacterial culture
The microbial strains used in this study were obtained from American Type Culture Collection (ATCC, Manassas, VA). The ATCC identification number for each strain was as follows: Escherichia coli JM109 (53323), E. coli O157:H7 (35150), vancomycin-susceptible Enterococcus faecalis (VSEF) (29212), vancomycin-resistant E. faecalis (VREF) (51299), Proteus mirabilis (29906), Pseudomonas aeruginosa (19143), methicillin-susceptible Staphylococcus aureus (MSSA) (29213), methicillin-resistant S. aureus (MRSA) (33591), and Staphylococcus epidermidis (35983). Experiments requiring transfer of biohazardous materials were conducted in a dedicated laminar flow hood equipped with UV lamp. Lyophilized bacteria were reconstituted in TSB and cultured overnight at 37 °C. A 1-mL aliquot of culture was grown in 100 mL of TSB for 2–4 h until reaching an optical density at 600 nm (OD600) ~ 0.15–0.3. The resulting culture was stored at −80 °C in 1-mL aliquots. For daily experiments, 1 mL of bacteria culture was grown in 100 mL of TSB overnight at 37 °C. Re-cultured in fresh TSB the next day, the bacteria were then grown to mid-exponential phase, as determined by OD600 measurements (approximately 1 × 108 CFU mL−1). The relationship between the OD600 and the concentration of bacteria in the culture suspension was calibrated for each strain using a Spectronic 301 spectrophotometer (Milton Roy, Ivyland, PA) and enumeration of colony forming units (cfu) from culture dilutions grown on TSA plates. For single-agent bactericidal assays conducted to determine the bactericidal activity of PROLI/NO, the 108 cfu mL−1 bacterial suspension was diluted 100-fold in TSB to obtain a final concentration of 1 × 106 cfu mL−1. For single-agent bactericidal assays employing AgSD and checkerboard assays, a 50-fold dilution in TSB was performed, resulting in a 2 × 106 cfu mL−1 bacterial concentration.
Single-agent bactericidal assays
The bactericidal efficacy of single agents (e.g., AgSD, PROLI/NO) after 120 min of exposure was evaluated against each pathogenic organism using a time-kill protocol. The minimum bactericidal concentration at 120 min (MBC120) was defined as the concentration of AgSD or PROLI/NO that resulted in a 3-log reduction in viability for a particular species over 120 min. Each strain of bacteria was tested in triplicate against 5 concentrations each of AgSD and PROLI/NO. To determine the efficacy of Ag+, solutions of AgSD in TSB were prepared and added to an equal volume of 2 × 106 cfu mL−1 bacterial suspension for a final starting innoculum concentration of 1 × 106 cfu mL−1. To evaluate the efficacy of NO, PROLI/NO was pre-weighed into chilled vials, and the appropriate volume of 1 × 106 cfu mL−1 bacterial suspension was added to obtain the target PROLI/NO concentration.
At each time point (0, 60, and 120 min), a 1:10 dilution of the microbial culture was prepared in PBS and a 100-μL aliquot of each dilution was spread onto TSA plates and incubated at 37 °C overnight. The number of colonies was enumerated to evaluate cell viability at 0, 60, and 120 min.
To circumvent common pitfalls associated with traditional efficacy techniques, some aspects of the bactericidal assays were modified from standard protocols.36 The most important features involved obtaining bactericidal (rather than inhibitory) concentrations and requiring efficacy over acute treatment windows (2 h). The rationale was to ensure swift and efficient bactericidal efficacy, as these therapeutic parameters discourage the selection of mutated resistant species. Additionally, we observed the susceptibility of a bacterial strain and its dose-response to an agent as a function of time by counting viable colonies at 60 and 120 min, rather than simply observing the all-or-none endpoint generated at 24 h by inhibitory (turbidity) determinations.
Checkerboard assay
The checkerboard method 33 was employed to experimentally determine the efficacy of AgSD and PROLI/NO in combination. Modifications analogous to those used in the single-agent bactericidal assays were adopted as described below. Briefly, bacteria (at a final innoculum concentration of 1 × 106 cfu mL−1) were incubated with an array of antimicrobial combinations of AgSD and PROLI/NO for 2 h at 37 °C. The highest concentration for each antimicrobial tested was a two-fold dilution of the concentration determined in the single-agent assay exhibiting rapid bactericidal activity (≥99%). Three additional dosages at stepwise, two-fold reductions in concentration were evaluated, resulting in 16 total combinations of AgSD and PROLI/NO tested against each strain of bacteria. For organisms that were particularly susceptible to the combination (i.e., MSSA, VSEF, VREF), lower concentrations of each agent were selected to probe the synergistic limit. Viable cells were enumerated at 0 and 120 min. The fractional bactericidal concentration index at 120 min (FBC120) was calculated using Equation 1:
| Eq. (1) |
adapted from the fractional inhibitory concentration index (FIC) reported by Elion et al., 37 where MBC120A and MBC120B are the values determined for agent A and B, respectively, in the single-agent assay; and, MBC120AB and MBC120BA are the concentrations of agent A and B that constituted the most effective bactericidal combination as determined by a 3-log reduction of viability. Synergy assays were conducted in three independent experiments for each strain of bacteria. A FBC120 < 0.5 was defined as synergistic, while a FBC120 < 0.25 was considered highly synergistic.
Results
Bactericidal activity of AgSD and PROLI/NO independently
The bactericidal activity of AgSD and PROLI/NO were evaluated against four Gram-negative (E. coli JM109, E. coli O157:H7, P. aeruginosa and P. mirabilis) and five Gram-positive (VSEF, VREF, MSSA, MRSA, and S. epidermidis) pathogenic strains of bacteria, including two antibiotic-resistant varieties. The concentrations required for bactericidal efficacy for AgSD against the panel of organisms spanned 3 orders of magnitude (Table 1). P. aeruginosa exhibited levels of susceptibility in the low micromolar range (56 μM, 0.020 g L−1) comparable to previous reports.4, 38 The bactericidal concentration for E. coli (0157:H7) was a full order of magnitude greater (560 μM, 0.20 g L−1) than P. aeruginosa, while MRSA required a ~five-fold greater dose (8960 μM, 3.20 g L−1) than the MSSA strain. Vancomycin-resistant Enterococcus faecalis required the greatest dose to achieve a 3-log reduction in viability (11200 μM, 4.0 g L−1).
TABLE 1.
Minimum bactericidal concentrations of PROLI/NO and AGSD required for a 3-log reduction in viable bacteria after 120 min of exposure.
| Species | Gram class | MBC120 AgSD (g L−1) | MBC120 PROLI/NO (g L−1) |
|---|---|---|---|
| E. faecalis | + | 2.00 | 72 |
| VREF | + | 4.00 | 60 |
| S. aureus | + | 0.600 | 48 |
| S. epidermidis | + | 0.100 | 36 |
| MRSA | + | 3.20 | 36 |
| E. coli (JM109) | − | 0.050 | 1 |
| E. coli (O157:H7) | − | 0.200 | 24 |
| P. aeruginosa | − | 0.020 | 8 |
| P. mirabilis | − | 0.100 | 12 |
Bactericidal concentrations of PROLI/NO also varied significantly between pathogens. The Gram-negative species E. coli (JM109) and P. aeruginosa required the lowest PROLI/NO doses at 1 g L−1 and 8 g L−1, respectively. The most susceptible Gram-positive strains were S. epidermidis and MRSA, each requiring 36 g L−1 for 3 logs of bactericidal activity (Table 1). VSEF exhibited the greatest tolerance to NO, withstanding concentrations of PROLI/NO up to 72 g L−1.
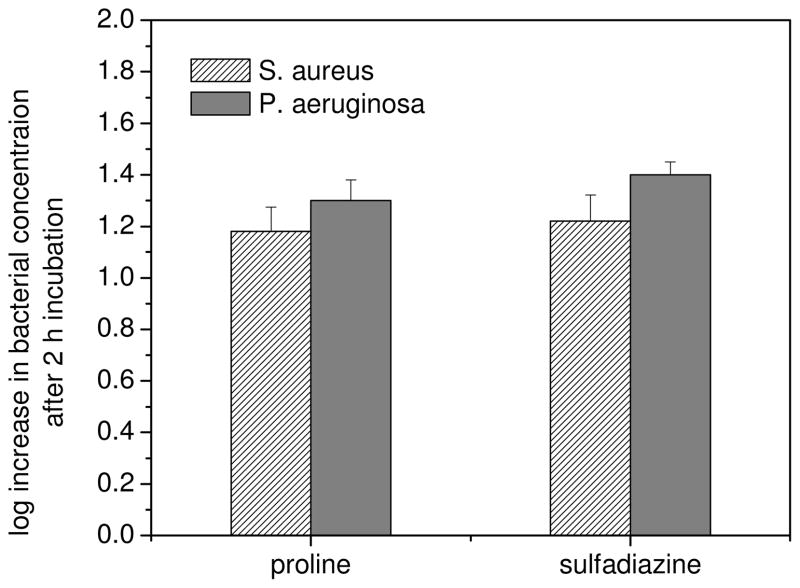
To verify that dissociated SD or regenerated proline did not contribute to observed cytotoxicity, we examined the bactericidal activity of these compounds with similar testing protocols. Molar concentrations of SD equivalent to the concentrations required for AgSD cytotoxicity against S. aureus and P. aeruginosa proved overwhelmingly nontoxic, having neither a static nor cidal effect. In all cases the bacteria continued to multiply during treatment, such that populations increased by more than 1 log over 2 h (Figure 1). Proline exhibited a similar non-toxic effect at concentrations equaling the MBC120 of PROLI/NO for the two species. Thus, the bactericidal activity of AgSD and PROLI/NO as single agents can be ascribed to Ag+ and NO, respectively, under the experimental protocol adopted in this study.
FIG. 1.
Change in concentration of S. aureus and P. aeruginosa after 2 h of exposure to proline and sulfadiazine at molar equivalents to the MBC of PROLI/NO and AgSD, respectively, in TSB at 37 °C.
The Gram-positive species studied generally exhibited superior tolerance to the single agents, Ag+ and NO, when compared to the Gram-negative species. Indeed, the two strains of E. faecalis indicated the greatest tolerance to both PROLI/NO and AgSD. The resilience of E. faecalis mirrors a report on the efficacy of dilute honey, another broad-spectrum antimicrobial, against a variety of bacterial species, where E. faecalis again demonstrated high levels of antimicrobial tolerance.39 Among the Gram-positive species examined in our study, the antibiotic-resistant ‘super bugs’ tended to demonstrate greater tolerance to AgSD, but similar tolerance to NO than their antibiotic-susceptible congeners. We observed that MRSA exhibited significantly greater tolerance to Ag+ than its methicillin-susceptible counterpart. Similarly, VREF required twice the dose of AgSD than the vancomycin-susceptible strain. The phenomenon that resistant bacteria selected by exposure to one antimicrobial frequently demonstrate resistance to other agents has been documented repeatedly.40–42 For instance, an AgSD-resistant strain of Enterbacter cloacae isolated from a burn wound unit also exhibited a resistance to kanamycin and carbenicillin not expressed by AgSD-susceptible strains.43
Synergistic activity of AgSD and PROLI/NO in combination
Measurement of the bactericidal endpoint over a short treatment duration (2 h) focused the definition of synergy in these experiments to short-term antimicrobial activity, as would be required for most topical applications of these drug combinations in vivo. Using the checkerboard technique, combinations of AgSD and PROLI/NO were screened against E. coli, VSEF, VREF, P. mirabilis, P. aeruginosa, MSSA, MRSA and S. epidermidis. Bactericidal synergism (FBC120 < 0.5) was evident for 4 out of 9 species tested. For one bacterial species, MRSA, the therapeutic combination was highly synergistic (FBC120 < 0.25). While Gram-positive bacteria were least susceptible to both AgSD and PROLI/NO as individual antimicrobial agents, the combination of AgSD and PROLI/NO was in fact synergistic against these same species, with the exception of S. epidermidis. For example, MRSA and both E. faecalis strains demonstrated high tolerance to each agent individually, but suffered the highest degree of susceptibility to the combination of AgSD and PROLI/NO, requiring only 6.25 – 12.5% of either agent in combination compared to the concentrations required for either of the agents alone. The MSSA strain also exhibited a high degree of susceptibility to this combination, although MRSA was affected to a greater degree. Of interest, P. aeruginosa was the most susceptible to AgSD alone, but synergy was not observed from the combination of AgSD and PROLI/NO.
Table 2 indicates the percent of each individual agent needed to elicit the greatest degree of synergy in combination. For the two most synergistic combinations (in Table 2, first two species listed), the dose of PROLI/NO was reduced to 6.25% values when combined with only 12.5% of the dose of AgSD. The much smaller percentage of PROLI/NO required for combined efficacy indicates the importance of PROLI/NO’s role in catalyzing the synergistic mechanism of action. Again using the checkerboard assay, the impact of varying the sequence of addition and spacing between agents was tested (Table 3). PROLI/NO (6 g L−1) and AgSD (0.200 g L−1) were added either simultaneously or at intervals of 15, 30, and 45 min using S. aureus as a test organism. The potential mechanistic implications of these studies are discussed below.
TABLE 2.
Typical concentrations of AgSD and PROLI/NO that elicited optimum synergistic effect, the correlating fold decrease over the respective values required for bactericidal activity alone, and the fractional bactericidal concentration (FBC120) for each species. The dashed line demarks the transition from synergistic to indifferent.
| Species | AgSD | PROLI/NO | |||
|---|---|---|---|---|---|
| combination (g L−1) | fraction (%) | combination (g L−1) | fraction (%) | FBC120 | |
| MRSA | 0.400 | 12.5 | 2.25 | 6.25 | 0.23 |
| VREF | 0.500 | 12.5 | 3.75 | 6.25 | 0.31 |
| E. faecalis | 0.250 | 12.5 | 9.0 | 12.5 | 0.33 |
| S. aureus | 0.100 | 16.7 | 12.0 | 25 | 0.42 |
| E. coli (O157:H7) | 0.050 | 25 | 6.0 | 25 | 0.53 |
| P. mirabilis | 0.050 | 50 | 0.75 | 6.25 | 0.56 |
| P. aeruginosa | 0.005 | 25 | 2.0 | 25 | 0.63 |
| E. coli (JM109) | 0.0125 | 25 | 0.25 | 12.5 | 0.67 |
| S. epidermidis | 0.050 | 50 | 18.0 | 50 | 1.00 |
TABLE 3.
Change in S. aureus viability at 120 min after exposure to 6 g L−1 PROLI/NO (PNO), 0.200 g L−1 AgSD, varying the sequence and interval between additions. Addition interval denotes time period between the addition of the first and second agent.
| Agent 1 | Addition interval (min) | Agent 2 | Bacterial viability change (log change) |
|---|---|---|---|
| AgSD | 45 | PNO | −1.61 |
| AgSD | 30 | PNO | −2.35 |
| AgSD | 15 | PNO | −2.64 |
| PNO | 45 | AgSD | −0.51 |
| PNO | 30 | AgSD | −0.37 |
| PNO | 15 | AgSD | −2.55 |
| AgSD | 0 | PNO | −2.46 |
| PNO | N/A | N/A | +0.11 |
| AgSD | N/A | N/A | −1.66 |
Discussion
Bactericidal efficacy of PROLI/NO and AgSD
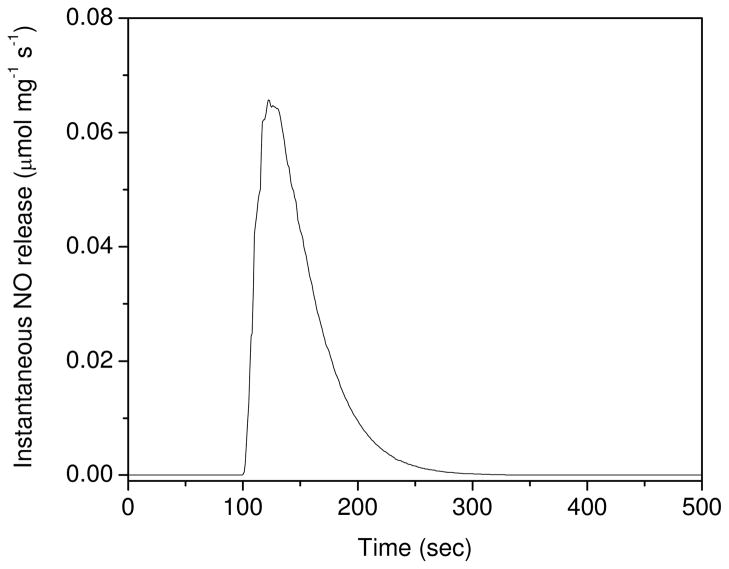
To understand the synergism observed in this study, it is first necessary to parse out the mechanisms of AgSD and PROLI/NO action. The various roles of NO in killing bacteria have been reviewed previously.10, 14, 17, 44 As an indiscriminate and short-lived reactant, the target sites available in the immediate vicinity of NO (i.e., thiols, amines, transition-metals, small molecules) play a key role in determining the type of antimicrobial action rendered. The NO donor used in our study (PROLI/NO) releases NO rapidly upon exposure to aqueous solution (t1/2 = 37 s, Figure 2),35, 45 where NO and its reactive intermediates likely modify biomolecules on the exterior of bacteria, particularly membrane-bound proteins and lipids. Reaction with O2− forms ONOO−, a strong oxidant that can degrade membranes through lipid peroxidation and oxidize nearby proteins, compromising cellular integrity. Additionally, NO is a lipophilic, uncharged, diatomic molecule that readily diffuses across lipid membranes where it may react with intercellular proteins and DNA, disrupting crucial cellular processes.. Thus, NO is both an extra- and intra-cellular threat to bacteria.
FIG. 2.
Instantaneous NO release from 0.133 mg PROLI/NO added to PBS (pH 7.4) at 37 °C.
Despite its broad application for treating chronic and burn wounds topically, AgSD’s mechanism(s) of action remains uncertain. In aqueous media, AgSD gradually dissociates into Ag+ and SD. Ionic silver reacts directly with thiol-containing amino acids.46 Mechanistic studies have shown that treatment with Ag+ affects DNA replication and cellular respiration, among other functions.1–6 A member of the sulfonamide family of semisynthetic antibacterial agents, SD is a biocide in its own right that can interfere with folate synthesis by competitively binding the enzyme dihydropteroate synthase within the cytoplasm of bacteria.47 This process is specifically detrimental to bacteria, as higher organisms obtain this metabolite through dietary ingestion. At physiological pH ~7 SD is negatively charged (pKa = 6.48)48 and less likely to diffuse across biological membranes to access its intracellular target.47 Despite the potential antimicrobial activity of SD, it is generally believed that Ag+ serves as the primary biocide upon AgSD dissociation, at least for topical applications.49 This phenomenon was verified in our experiments by showing that treatment with SD alone at concentrations equivalent to the MBC of AgSD did not affect the viability of any of the organisms studied (Figure 1).
As evidenced by the ineffectiveness of SD, the ability of a biocide to access target sites is imperative for antimicrobial efficacy. The outer membrane characteristic of Gram-negative species acts as a particularly efficient permeability barrier, conferring intrinsic resistance to host defense mechanisms, bile salts and digestive enzymes, and many biocides that are effective against other types of bacteria.50 In the absence of differences in specificity, Gram-negative bacteria frequently exhibit improved tolerance to agents that function intracellularly compared to Gram-positive species.51 Hence, the greater efficacy of Ag+ and NO (individually) observed against Gram-negative species may indicate that important targets of these biocides reside on the exterior of the bacteria where the rate of passive diffusion is less important. When considering the reactivity of Ag+, NO, and NO-derived by-products, this is not entirely surprising. Empirical evidence drawn from morphological observation supports this hypothesis. For example, electron micrographs of AgSD-treated P. aeruginosa and Enterobacter cloacae have indicated altered cell wall morphology, while resistant species did not show any changes.43, 52
Proposed mechanisms for the synergistic action of PROLI/NO and AgSD
Our control experiments clearly indicate that Ag+ and NO are the bactericidal agents eliciting synergy. Two possible cooperative mechanisms seem plausible based on the cumulative knowledge of Ag+ and NO activity, neither of which are mutually exclusive. Agents that act by disrupting the structure of the lipid bilayer or otherwise compromising the cell wall, independent of the level of individual bactericidal activity, should in principle work synergistically when combined with a second agent whose activity is frustrated by low levels of permeability. We have previously reported that NO is a potent membrane degradation agent, damaging the bacterial membrane of gram-negative species. Pseudomonas aeruginosa treated with NO were imaged with atomic force microscopy, revealing widespread structural deformation and collapse of the bacterial membrane.45 These results were further corroborated by the observed penetration of propidium iodide, a fluorescent dye that will only penetrate compromised cell membranes, upon exposure of P. aeruginosa to NO.24 Synergistic mechanisms between cell wall- and cytoplasm-active agents have been repeatedly demonstrated (e.g., the improved permeability of streptomycin into E. faecalis in concert with cell wall-active agents such as penicillin and vancomycin),53, 54 and a similar phenomenon has been shown against other Gram-negative and -positive species.34 We hypothesize that such synergism may occur if cell wall or membrane damage elicited by Ag+ and, particularly, NO significantly increases the permeability of the bacteria cell wall. Both of the antimicrobial agents evaluated in our study have intracellular activity that would be expected to be enhanced by a faster rate of entry into the bacteria. Furthermore, increased permeability of the lipid membrane would improve intracellular access for anionic SD compounds.
Among other bactericidal mechanisms, Ag+ has been implicated in disruption of cellular respiration. The uncoupling of the respiratory chain initially stimulates respiration as bacteria attempt to regenerate the declining proton gradient across the membrane. Holt and Bard showed that AgNO3 inhibited the respiratory chain by preventing the transport of protons outside of the cell.6 One important implication of their study was the probable accumulation of reactive oxygen species such as O2− and OH− at the membrane. The bactericidal efficacy of NO would increase with the number of reactive by-products produced (i.e., peroxynitrite).
The underlying synergistic mechanism may be indirectly probed by altering the sequence of addition. If decoupling of cellular respiration plays an important role in the synergistic mechanism, then the action of Ag+ must precede that of the highly reactive NO produced from rapidly decomposing PROLI/NO unless increased cellular permeability is the major contributing factor to the synergistic activity. In the latter case, the PROLI/NO likely elicits the degradation of membrane lipids prior to the subsequent contribution from increased intracellular access of both Ag+ and NO. A series of bactericidal assays were thus performed varying both the order and time of addition for each bactericidal agent (Table 3). The addition of PROLI/NO prior to AgSD within a 15 min window and AgSD prior to PROLI/NO within a 30 min window resulted in the greatest synergistic effect, suggesting that the order of addition is not important within a narrow time period. Antimicrobial activity was diminished when AgSD was added >30 min prior to PROLI/NO, perhaps due to reduced potency of the AgSD in solution before any action on the cell walls by PROLIN/NO). Similarly, antimicrobial efficacy was reduced when PROLI/NO was added >15 min prior to AgSD, likely due to the short half-life of NO at physiological pH (~40 s). We hypothesize that at additional periods beyond 15 min for PROLI/NO and 30 min for AgSD, either cell division may result in new, non-damaged cells, or cell wall repair occurs despite the short-lived presence of sub-bactericidal doses of NO.55 Any new or repaired cells would then be unaffected by the addition of sub-bactericidal doses of AgSD, allowing normal proliferation to occur.
To evaluate the potential effect of increased SD permeability, S. aureus bacteria were treated with a range of SD concentrations in combination with doses of PROLI/NO at one half and one quarter of the respective bactericidal levels. No enhanced activity was observed for S. aureus, indicating that SD is not involved in the primary synergistic mechanism. Even the lowest concentration of SD evaluated (0.105 g L−1) was almost three-fold greater than the equivalent concentration of AgSD required for maximum cooperativity against P. aeruginosa (0.005 g L−1) with a 12 g L−1 dose of PROLI/NO. These results indicated that combination treatment with NO and SD was not particularly effective against S. aureus over a 2 h time frame, although some enhanced effect may be expected to occur over longer periods as endogenous folate reserves are depleted and cellular activity decreases due to starvation. The primary mechanism of the acute synergy observed at 2 h involves both NO and Ag+, and the evidence revealed in this study indicates that increased intracellular concentrations of these two agents following membrane damage may play a significant role in the observed synergistic activity.
Conclusions
The combination of AgSD and PROLI/NO is synergistic across a wide range of Gram-positive bacteria, including antibiotic-resistant ‘super bugs’. By varying the interval of addition of AgSD and PROLI/NO, we demonstrated that the duration between dosing of individual agents is important in eliciting maximum synergistic activity. When the concentration of AgSD or PROLI/NO is sub-bactericidal, the delay in the addition of the second agent should not exceed some threshold time limit, presumably as the bacteria are capable of repairing the membrane damage or dividing to form healthy cells, even while under stress.
When AgSD and PROLI/NO were evaluated against bacteria individually, Gram-negative bacteria were the most susceptible to treatment, with as little as 0.020 g L−1 AgSD and 1 g L−1 PROLI/NO required for a 3-log reduction in viability of P. aeruginosa and E. coli (JM109), respectively. Gram-positive bacteria were the most susceptible to the cooperative effect of the combination of AgSD and PROLI/NO. In the case of MRSA, only 12.5% of the AgSD and 6.25% of the PROLI/NO was required to be bactericidal in combination compared to the concentrations needed when used individually. The evidence presented herein provides an impetus to further investigate the clinical uses of NO in combination with AgSD and other antibiotics. Possessing the ability to rapidly eradicate a wide range of bacterial species, such combinations may be particularly useful in topical wound treatments due to the potential for treating polymicrobial and antibiotic-resistant infections while reducing selection for resistant species. Further mechanistic studies aimed at elucidating the synergistic action of AgSD and NO would provide a strong foundation for developing and improving additional combination therapies. In addition, studies examining both the cytotoxicity and in vivo efficacy of the therapy are necessary prior to clinical application.
Acknowledgments
This work was supported by the National Institute of Health (NIH EB000708).
References
- 1.Russell AD, Hugo WB. Antimicrobial activity and action of silver. Prog Med Chem. 1994;31:351–371. doi: 10.1016/s0079-6468(08)70024-9. [DOI] [PubMed] [Google Scholar]
- 2.Bragg PD, Rainne DG. The effect of silver ions on the respiratory chain of Escherichia coli. Can J Microbiol. 1974;20:883–889. doi: 10.1139/m74-135. [DOI] [PubMed] [Google Scholar]
- 3.Dibrov P, Dzioba J, Gosink KK, Häse CC. Chemiosmotic mechanism of antimicrobial activity of Ag+ in Vibrio cholerae. Antimicrob Agents Chemother. 2002;46:2668–2670. doi: 10.1128/AAC.46.8.2668-2670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modak SM, Fox J, CL Binding of silver sulfadiazine to the cellular components of Pseudomonas aeruginosa. Biochem Pharmac. 1973;22:2391–2404. doi: 10.1016/0006-2952(73)90341-9. [DOI] [PubMed] [Google Scholar]
- 5.Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res. 2000;52:662–668. doi: 10.1002/1097-4636(20001215)52:4<662::aid-jbm10>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Holt KB, Bard AJ. Interaction of silver (I) ions with the respiratory chain of Escherichia coli: An electrochemical and scanning electrochemical microscopy study of the antimicrobial mechanism of micromolar Ag+ Biochemistry. 2005;44:13214–13223. doi: 10.1021/bi0508542. [DOI] [PubMed] [Google Scholar]
- 7.Silver S, Phung LT, Silver G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J Ind Microbiol Biotechnol. 2006;33:627–634. doi: 10.1007/s10295-006-0139-7. [DOI] [PubMed] [Google Scholar]
- 8.Carr HS, Wlodkowski TJ, Rosenkranz HS. Silver sulfadiazine: in vitro antibacterial activity. Antimicrob Agents Chemother. 1973;4:585–587. doi: 10.1128/aac.4.5.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wlodkowski TJ, Rosenkranz HS. Antifungal activity of silver sulphadiazine. Lancet. 1973;ii:739–740. doi: 10.1016/s0140-6736(73)92576-2. [DOI] [PubMed] [Google Scholar]
- 10.Fang FC. Host/pathogen interactions: Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 12.Foley E, O’Farrell PH. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 2003;17:115–125. doi: 10.1101/gad.1018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 14.Mannick JB. Immunoregulatory and antimicrobial effects of nitrogen oxides. Proc Am Thorac Soc. 2006;3:161–165. doi: 10.1513/pats.200505-048BG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wink DA, Mitchell JB. Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol and Med. 1998;25(4):434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 16.Moller MN, Li Q, Lancaster JRJ, Denicola A. Acceleration of nitric oxide autooxidation and nitrosation by membranes. Life. 2007;59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 17.Zaki MH, Akuta T, Akaike T. Nitric oxide-induced nitrative stress involved in microbial pathogenesis. J Pharmacol Sci. 2005;98:117–129. doi: 10.1254/jphs.crj05004x. [DOI] [PubMed] [Google Scholar]
- 18.Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, Dunams TM, Cebula TA, Koch WH, Andrews AW, Allen JS, Keefer LK. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 19.Schapiro JM, Libby SJ, Fang FC. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. PNAS. 2003;100:8496–8501. doi: 10.1073/pnas.1033133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghaffari A, Miller CC, McMullin B, Ghahary A. Potential application of gaseous nitric oxide as a topical antimicrobial agent. Nitric Oxide. 2006;14:21–29. doi: 10.1016/j.niox.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Raulli R, McElhaney-Feser G, Hrabie JA, Cihlar RL. Antimicrobial properties of nitric oxide using diazeniumdiolates as the nitric oxide donor. Rec Res Devel Microbiology. 2002;6:177–183. [Google Scholar]
- 22.Keefer LK. Progress toward clinical application of the nitric oxide-releasing diazeniumdiolates. Annu Rev Pharmacol Toxicol. 2003;43:585–607. doi: 10.1146/annurev.pharmtox.43.100901.135831. [DOI] [PubMed] [Google Scholar]
- 23.Hetrick EM, Schoenfisch MH. Antibacterial nitric oxide-releasing xerogels: Cell viability and parallel plate flow cell adhesion studies. Biomaterials. 2007;28:1948–1956. doi: 10.1016/j.biomaterials.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Hetrick EM, Shin JH, Stasko NA, Johnson BA, Wespe DA, Holmuhamedov E, Schoenfisch MH. Bactericidal efficacy of nitric oxide-releasing silica nanoparticles. ACS Nano. 2007;2:235–246. doi: 10.1021/nn700191f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nablo BJ, Rothrock AR, Schoenfisch MH. Nitric oxide-releasing sol-gels as antibacterial coatings for orthopedic implants. Biomaterials. 2005;26:917–924. doi: 10.1016/j.biomaterials.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 26.Levy SB. The Antibiotic Paradox. 2. Perseus; Cambridge: 2002. [Google Scholar]
- 27.Kliebe C, Nies BA, Meyer JF, Tolxdorff-Neutzling RM, Wiedemann B. Evolution of plasmid-encoded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985;28:302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall RM, Collins CM. Mobile gene cassettes and ntegrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 29.Levy SB. Multidrug resistance: a sign of the times. New Eng J Med. 1998;338:1376–1378. doi: 10.1056/NEJM199805073381909. [DOI] [PubMed] [Google Scholar]
- 30.Hiramatsu K. Vancomycin resistance in Staphylococci. Drug Res Updates. 1998;1:135–150. doi: 10.1016/s1368-7646(98)80029-0. [DOI] [PubMed] [Google Scholar]
- 31.Michel M, Gutmann L. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: therapeutic realities and possibilities. Lancet. 1997;349:1901–1906. doi: 10.1016/s0140-6736(96)11192-2. [DOI] [PubMed] [Google Scholar]
- 32.Cottarel G, Wierzbowski J. Combination drugs, an emerging option for antibacterial therapy. Trends in Biotechnology. 2007;25:547–555. doi: 10.1016/j.tibtech.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Eliopoulos GM, Moellering JRC. Antimicrobial Combinations. In: Lorian V, editor. Antibiotics in Laboratory Medicine. 4. Williams & Wilkins; Baltimore: 1996. pp. 330–396. [Google Scholar]
- 34.Eliopoulos GM, Eliopoulos CT. Antibiotic combinations: Should they be tested? Clin Microbiol Rev. 1988;1(2):139–156. doi: 10.1128/cmr.1.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saavedra JE, Southan GJ, Davies KM, Lundell A, Markou C, Hanson SR, Adrie C, Hurford WE, Zapol WM, Keefer LK. Localizing antithrombotic and vasodilatory activity with a novel, ultrafast nitric oxide donor. J Med Chem. 1996;39:4361–4365. doi: 10.1021/jm960616s. [DOI] [PubMed] [Google Scholar]
- 36.Amsterdam D. Susceptibility Testing of Antimicrobials in Liquid Media. In: Lorian V, editor. Antibiotics in Laboratory Medicine. 4. Williams & Wilkins; Baltimore: 1996. pp. 52–111. [Google Scholar]
- 37.Elion GB, Singer S, Hitchings CH. Antagonists of nucleic acid derivitives. VIII. Synergism in combinations of biochemically related antimetabolites. J Biol Chem. 1954;208:477–488. [PubMed] [Google Scholar]
- 38.Rosenkranz HS, Carr HS. Silver sulfadiazine: Effect on the growth and metabolism of bacteria. Antimicrob Agents Chemother. 1972;2:367–372. doi: 10.1128/aac.2.5.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwakman PHS, Van den Akker JPC, Güçlü A, Aslami H, Binnekade JM, de Boer L, Boszhard L, Paulus F, Middelhoek P, te Velde AA, Vandenbroucke-Grauls CMJE, Schultz MJ. Medical-grade honey kills antibiotic-resistant bacteria in vitro and eradicates skin colonization. Clin Infect Dis. 2008;46:1677–1682. doi: 10.1086/587892. [DOI] [PubMed] [Google Scholar]
- 40.Akimitsu N, Hamamoto H, Inoue R, Shoji M, Akamine A, Takemori K, Hamasaki N, Sekimizu K. Increase in resistance of methicillin-resistant Staphylococcus aureus to β-lactams caused by mutations conferring resistance to benzalkonium chloride, a disinfectant widely used in hospitals. Antimicrob Agents Chemother. 1999;43:3042–3043. doi: 10.1128/aac.43.12.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta N, Faiers MC, Reeves DS, Brumfitt W, Orskov F, Orskov I. R factors in Escherichia coli in faeces after oral chemotherapy in general practice. Lancet. 1971;1:312–315. doi: 10.1016/s0140-6736(71)91042-7. [DOI] [PubMed] [Google Scholar]
- 42.Levy SB, Marshall B, Schluederberg S, Rowse D, Davis J. High frequency of antimicrobial resistance in human fecal flora. Antimicrob Agents Chemother. 1988;32:1801–1806. doi: 10.1128/aac.32.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenkranz HS, Coward JE, Wlodkowski TJ, Carr HS. Properties of silver sulfadiazine-resistant Enterobacter cloacae. Antimicrob Agents Chemother. 1973;5:199–201. doi: 10.1128/aac.5.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Groote MA, Fang FC. NO inhibitions: antimicrobial properties of nitric oxide. Clin Infect Dis. 1995;21 (Suppl 2):S162–S165. doi: 10.1093/clinids/21.supplement_2.s162. [DOI] [PubMed] [Google Scholar]
- 45.Deupree SM, Schoenfisch MH. Morphological analysis of the antimicrobial action of nitric oxide on Gram-negative pathogens using atomic force microscopy. Acta Biomater. 2009 doi: 10.1016/j.actbio.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liau SY, Read DC, Pugh WJ, Furr JR, Russell AD. Interaction of silver nitrate with readily identifiable groups: relationship to the antibacterial action of silver ions. Letters in Applied Microbiology. 1997;25:279–283. doi: 10.1046/j.1472-765x.1997.00219.x. [DOI] [PubMed] [Google Scholar]
- 47.Zarfl C, Matthies M, Klasmeier J. A mechanistical model for the uptake of sulfonamides by bacteria. Chemosphere. 2008;70:753–760. doi: 10.1016/j.chemosphere.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 48.Sakurai H, Ishimitsu T. Microionization constants of sulphonamides. Talanta. 1980;27:293–298. doi: 10.1016/0039-9140(80)80061-0. [DOI] [PubMed] [Google Scholar]
- 49.Klasen HJ. Historical review of the use of silver in the treatment of burns. I. Early uses. Burns. 2000;26:117–130. doi: 10.1016/s0305-4179(99)00108-4. [DOI] [PubMed] [Google Scholar]
- 50.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiological reviews. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maillard J-Y. Bacterial target sites for biocide action. Journal of Applied Microbiology. 2002;92(s1):16S–27S. [PubMed] [Google Scholar]
- 52.Coward JE, Carr HS, Rosenkranz HS. Silver sulfadiazine: Effect on the ultrastructure of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1973;3:621–624. doi: 10.1128/aac.3.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moellering JRC, Weinberg AN. Studies on antibiotic synergism against enterococci. II. Effect of various antibiotics on the uptake of 14C-labeled streptomycin by enterococci. J Clin Invest. 1971;50:2580–2584. doi: 10.1172/JCI106758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moellering JRC, Wennersten C, Weinberg AN. Studies on antibiotic synergism against enterococci. I. Bacteriologic studies. J Lab Clin Med. 1971;77:821–828. [PubMed] [Google Scholar]
- 55.Chilton P, Isaacs NS, Manas P, Mackey BM. Biosynthetic requirements for the repair of membrane damage in pressure-treated Escherichia coli. International Journal of Food Microbiology. 2001;71(1):101–104. doi: 10.1016/s0168-1605(01)00566-9. [DOI] [PubMed] [Google Scholar]