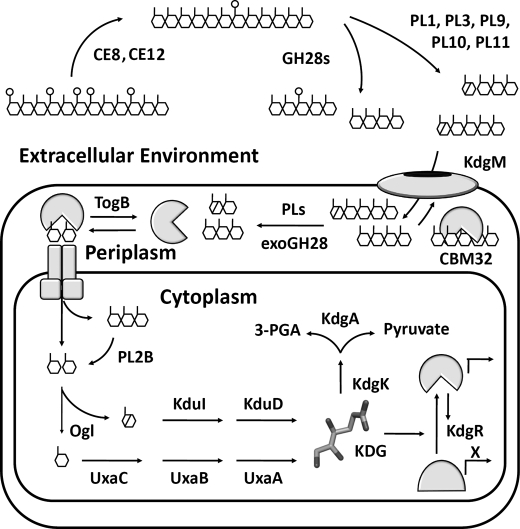
FIGURE 1.
General pectin degradation pathway found in pectinolytic bacteria from Enterobacteraciae (11). Extracellular pectin depolymerization is primarily restricted to phytopathogens and involves a consortium of enzymes that liberate fragments from the highly polymerized pectic network within the plant cell wall. Pectinases involved in these processes include PLs, polygalacturonases (GH28s), and carbohydrate esterases (CE8). Products diffuse into the periplasmic space through anionic porins belonging to the KdgM family, where a specialized collection of proteins operates to retain and concentrate (CBM32) and process pectic substrates into mono-, di-, and trisaccharides (PLs and exoGH28). Intracellular transport of oligogalacturonides is facilitated by integral membrane systems, including the ATP-dependent transporter TogMNAB. Within the cell, pectin catabolism remains poorly understood at the structural level; however, a pathway has been established based upon genetics, biochemistry, and comparative genomics (7, 12). There are two branches of metabolism dedicated to saturated monogalacturonate (GalA; UxaA, UxaB, and UxaC), and unsaturated monogalacturonate (DKI; KduI and KduD), the product of pectate lyases. These pathways converge to produce KDG, which is the inducer of KdgR, a repressor protein that regulates the expression of virtually every pectin utilization gene known. KDG is converted into the metabolites 3-phosphoglyceraldehyde and pyruvate by a two-step phosphorylation and hydrolysis reaction path catalyzed by KdgK and KdgA, respectively.