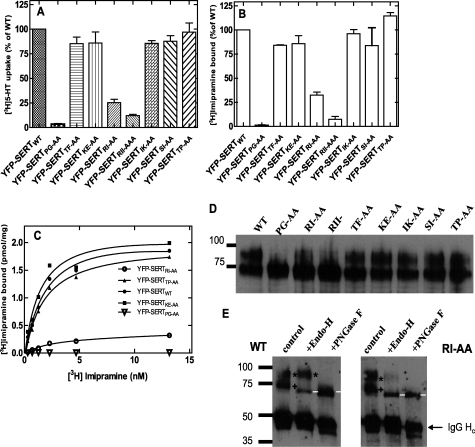
FIGURE 4.
Mutations of SERT in the position PG601,602, RI607,608, and RII607–609 blunt 5-HT uptake and binding of [3H]imipramine. HEK293 cells were transiently transfected with plasmids encoding the indicated mutants. A, cellular uptake of [3H]5-HT was determined at 3 μm [3H]5-HT; assay conditions were otherwise as outlined in the legend to Fig. 2. All transfections and determinations were done in parallel. To account for interassay variation in transfection efficiency, data were normalized to uptake determined in wild type SERT expressing cells; this 100% uptake corresponded to 27.5 ± 1.9 pmol/106 cells/min. B, binding of [3H]imipramine (3.6 nm) to membranes prepared from cells transiently expressing different versions of SERT; this 100% binding corresponded to 1.34 ± 0.03 pmol/mg. Data in panels A and B are means from five independent experiments carried out in duplicate; error bars indicate S.E. C, binding of [3H]imipramine to membranes (15–30 μg/assay) prepared from cells transiently expressing wild-type SERT, SERT-PG601,602AA, SERT-KE605–606AA, SERT-RI607,608AA, and SERT-TP613–614AA. Data are from a representative experiment done in duplicate which is representative for at least two additional experiments (see also supplemental Table S2). D, membranes were prepared from transiently transfected cells; the YFP-tagged versions of SERT were visualized with an anti-GFP antibody. E, wild-type SERT and SERT-RI607,608AA were enriched by immunoprecipitation, denatured, and subjected to deglycosylation by incubation (for 1 h at 37 °C) in the absence (control) and presence of endoglycosidase H (Endo H) or PNGase F. * denotes the mature glycosylated upper band, which is resistant to Endo H; + is the lower band, which is cleaved by Endo H to generate a smaller product (marked with white line). Note that the immunoglobulin heavy chain is also visible (IgG Hc) and that this is deglycosylated by PNGase F. Numbers indicate the position of molecular mass markers (in kDa).