Abstract
Piwi proteins and their partner small RNAs play an essential role in fertility, germ-line stem cell development, and the basic control and evolution of animal genomes. However, little knowledge exists regarding piRNA biogenesis. Utilizing microfluidic chip analysis, we present a quantitative profile of zebrafish piRNAs expressed differentially between testis and ovary. The sex-specific piRNAs are derived from separate loci of repeat elements in the genome. Ovarian piRNAs can be categorized into groups that reach up to 92 members, indicating a sex-specific arrangement of piRNA genes in the genome. Furthermore, precursor piRNAs preferentially form a hairpin structure at the 3′end, which seem to favor the generation of mature sex-specific piRNAs. In addition, the mature piRNAs from both the testis and the ovary are 2′-O-methylated at their 3′ ends.
SMALL RNAs, ranging from 19 to 30 nucleotides (nt) in length, constitute a large family of regulatory molecules with diverse functions in invertebrates, vertebrates, plants, and fungi (Bartel 2004; Nakayashiki 2005). Two major classes of small RNAs are microRNAs (miRNAs) and small interfering RNAs (siRNAs). The functions of small RNAs have been conserved through evolution; they have been shown to inhibit gene expression at the levels of mRNA degradation, translational repression, chromatin modification, heterochromatin formation, and DNA elimination (Mochizuki et al. 2002; Bartel 2004; Kim et al. 2005; Brodersen and Voinnet 2006; Lee and Collins 2006; Vaucheret 2006).
Over the past few years, focus on the genetics of small RNAs has helped clarify the mechanisms behind the regulation of these molecules. While hundreds of small RNAs have been identified from mammalian somatic tissues, relatively little is known about small RNAs in germ cells. A recent breakthrough has been the identification of small RNAs that associate with Piwi proteins (piRNAs) from Drosophila and mammalian gonads (Aravin et al. 2001, 2006; Girard et al. 2006; Grivna et al. 2006; Vagin et al. 2006; Watanabe et al. 2006). piRNAs and their interacting proteins Ziwi/Zili have also been identified in zebrafish (Houwing et al. 2007, 2008). Increasing evidence indicates that piRNAs play roles mainly in germ cell differentiation and genomic stability (Carthew 2006; Lau et al. 2006; Vagin et al. 2006; Brennecke et al. 2007; Chambeyron et al. 2008; Klattenhoff and Theurkauf 2008; Kuramochi-Miyagawa et al. 2008; Kim et al. 2009; Lim et al. 2009; Unhavaithaya et al. 2009). Moreover, although piRNAs are mostly expressed in germ line cells, recent studies showed piRNA expression in nongerm cells, for example, T-cell lines (Jurkat cells and MT4) (Azuma-Mukai et al. 2008; Yeung et al. 2009), indicating other functions such as in the immune system. piRNAs do not appear to be derived from double-stranded RNA precursors, and their biogenesis mechanisms, although unclear, may be distinct from those of siRNA and miRNA. Recently, two distinct piRNA production pathways were further proposed: the “ping-pong” model (Brennecke et al. 2007; Gunawardane et al. 2007) and the Ago3-independent piRNA pathway centered on Piwi in somatic cells (Li et al. 2009; Malone et al. 2009). However, the mechanistic pathways of piRNA activity and their biogenesis are still largely unknown.
Teleost fishes comprise >24,000 species, accounting for more than half of extant vertebrate species, displaying remarkable variation in morphological and physiological adaptations (see review in Zhou et al. 2001). Recently, Houwing et al. (2007, 2008) reported findings on Ziwi/Zili and associated piRNAs, implicating roles in germ cell differentiation, meiosis, and transposon silencing in the germline of the zebrafish. However, some of the identified zebrafish piRNAs are nonrepetitive and nontransposon-related piRNAs, suggesting that piRNAs may have additional unknown roles. In this study, we show that for males and females, piRNAs are specifically derived from separate loci of the repeat elements, and that ovarian piRNAs are far more often associated in groups. Genomic analysis of piRNAs indicates a tendency to folding at the 3′ end of the piRNA precursor, which may favor cleavage of the piRNA precursor to generate mature sex-specific piRNAs. Furthermore, methylation modification occurs at the 2′-O-hydroxyl group on the ribose of the final 3′ nucleotide in both the testis and the ovary.
MATERIALS AND METHODS
piRNA cloning:
Total RNAs of zebrafish ovary were isolated using Trizol (Invitrogen, Carlsbad, CA), separated on a 15% denaturing PAGE gel and visualized by SYBR-gold (Invitrogen) staining. Small RNAs of ∼30 nt were cut and purified from PAGE gel. Small RNAs were ligated to a 3′ adapter (5′AppCTGTAGGCACCATCAddA3′) and 5′ acceptor (5′ATCGTaggcaccugaaa3′, lowercase RNA) sequentially and then RT–PCR amplified as described (Lau et al. 2001). The PCR products were digested by BanI and concatamerized. The concatamers were cloned into a TOPO vector according to the manufacturer's protocol (Invitrogen) and sequenced by ABI 3730 autosequencer. Sequences of small RNAs ∼30 nt were identified by presence of the adapter sequences. Usually, each clone contains ∼3–4 small RNA reads.
Pare-piRNA and intermediate (noncoding RNA) cloning:
Small RNAs of zebrafish testis were extracted using the MicroRNA extraction kit (BioTeke, Beijing, China), separated on a 15% denaturing PAGE gel, and visualized by SYBR-gold (Invitrogen) staining. Around 40–60 nt of small RNAs from testis were cut, purified, and ligated to a 3′ adapter (5′AppCTGTAGGCACCATCAddA3′). Reverse transcription was performed with the RT primer (5′ATTGATGGTGCCTAC3′), and pre-piRNA sequences were amplified using the forward primer (5′TGCAAGTCCAGGG3′) and the reverse primer (5′ATTGATGGTGCCTACAG3′). The PCR products were cloned into a pGEM-T Easy vector (Promega, Madison, WI) and sequenced.
Genome mapping of cloned piRNAs:
Beginning with raw reads of the cloned sequence, we wrote a Java program to recognize the adapters and extract the small RNA sequences. piRNAs were mapped to a Zv6 assembly of the zebrafish genome using the University of California Santa Cruz (UCSC)'s Blat search (http://genome.ucsc.edu/cgi-bin/hgTables?org=Zebrafish&db=danRer5). For each piRNA sequence, we used only the best matches up to a maximum of two differences including mismatch, insertion, and deletion. All data (sequences, mapping information) were entered into a relational database. We used these genome locations to obtain genome annotations of piRNAs from the tracks in UCSC with the tool “Table” (http://genome.ucsc.edu/cgi-bin/hgTables?org=Zebrafish&db=danRer5).
Functional annotation:
We assembled a database of RNAs with known function such as rRNA, tRNA, microRNA, snoRNA, snRNA, and mRNA. We obtained part of the sequences by querying GenBank with the appropriate feature key. The other sequence data were downloaded from the NONCODE database (noncoding RNAs, http://noncode.bioinfo.org.cn), the snoRNA database (http://www-snorna.biotoul.fr), and the microRNA database (ftp://ftp.sanger.ac.uk/pub/mirbase/sequences/CURRENT). We used the RepeatMasker (http://www.repeatmasker.org/) results from UCSC for repeat annotation. All small RNAs obtained by cloning were compared with functionally annotated sequences using an in-house BLAST program. For each small RNA sequence, the best alignments to a functionally annotated sequence (up to two errors at most) were used to assign a functional category to the small RNA.
Secondary structure analysis:
We used the genome location of piRNAs or miRNAs to extract 400-nt sequences, with 200 nt upstream and 200 nt downstream of the 5′ or 3′ end of piRNA or miRNAs. Mfold software (http://mfold.bioinfo.rpi.edu/, version 3.2) was used to predict the secondary structure of the extracted regions. The minimum free energy structure was used to determine an average profile of paired nucleotides along the sequence, on the basis of the methods described (Aravin et al. 2006). The fraction of paired bases of each position along the sequence was calculated using the equation Vi = Mi/N (Vi, fraction of paired bases at the position i; Mi, paired numbers at the position i for all the minimum free energy structures; N, total numbers of the sequences). For piRNA folding analysis, we further calculated the 6-nt paired sequence (4–6 paired bases were defined as a paired sequence within the 6-nt sequence) numbers at each position along the piRNA sequence by scanning and comparison with the up- or downstream 200 nt sequences, on the basis of the folding structure with minimum free energy.
Microarray:
Two identical sets of microfluidic chips, each consisting of 3466 piRNAs, which were selected randomly from the piRNA sequence pool we cloned, were made by LC Sciences, Houston, TX. Microarray assay was performed using a service provider (LC Sciences). The assay started from 2 to 5 μg of the total RNA sample, which was size fractionated using a YM-100 Microcon centrifugal filter (Millipore, Billerica, MA), and the small RNAs (<300 nt) isolated were 3′ extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining. Two different tags were used for the two RNA samples in dual-sample experiments. Hybridization was performed overnight on a μParaflo microfluidic chip using a microcirculation pump (Atactic Technologies). The detection probes were made by in situ synthesis using photogenerated reagent (PGR) chemistry. Hybridization required 100 μl of 6× SSPE buffer containing 25% formamide at 34°. After hybridization, detection involved fluorescence labeling using tag-specific Cy3 and Cy5 dyes. Hybridization images were collected using a laser scanner (GenePix 4000B, Molecular Devices) and digitized using Array-Pro image analysis software (Media Cybernetics). Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter (locally weighted regression). For two-color experiments, the ratio of the two sets of detected signals (log2 transformed, balanced) and P-values of the t-tests were calculated. Differentially detected signals were those with P-values <0.01.
Northern blotting hybridization:
Small RNAs of zebrafish ovary and testis were extracted using a MicroRNA extraction kit (BioTeke), visualized by SYBR-gold (Invitrogen) staining, and electroblotted onto IMMOBILON-NY+ membranes (Millipore). The membranes were UV crosslinked and hybridized at 42° overnight with 32P end-labeled oligonucleotide probes in UltraHyb (Ambion, Austin, TX). Probes for detecting piRNAs were complementary to the piRNA sequences. The degenerate probe for group 1 piRNAs was 5′GHCCACCWMTCTCCCTGGACTTGCA3′ and for group 11 piRNAs, it was 5′GCCTYTSACCCCRGCAAGCTACCCA3′. After hybridization, membranes were washed with 2× SSC containing 0.5% SDS at 42° three times, 15 min each. The membranes were exposed to phosphor screens that were scanned using a variable scanner, the Typhoon 9200 (Amersham Pharmacia Biotech, Uppsala, Sweden).
2′-O-methylation analysis:
Zebrafish piRNAs (∼5 pmol) were digested with RNase T2 into 3′-, 5′-diphospho nucleotides, 3′-monophospho nucleotides, and 3′-terminal nucleosides at 37° for 3 hr in 5 μl of reaction mixture containing 10 mm ammonium acetate (pH 5.3) and 1.25 U/ml RNase T2 (GE Healthcare) as described (Ohara et al. 2007; Suzuki et al. 2007). The reaction solution was evaporated and dissolved in 1.5 μl of 50 mm ammonium acetate (pH 5.3). The procedure for liquid chromatography/mass spectrometry (LC/MS) analysis has been described (Ikeuchi et al. 2006). To analyze a limited quantity of nucleosides, we devised a system of capillary LC coupled with electrospray ionization mass spectrometry. A QSTAR XL Hybrid LC/MS/MS system (Applied Biosystems) equipped with a NANOSPRAY II nanoelectrospray (Applied Biosystems) source, a Nanovolume Valve (Valco Instruments), and a splitless nano HPLC system DiNa (KYA Technologies) was used to analyze RNase T2-digested piRNAs. The digest was loaded onto an ODS capillary column directly via a 1-μl loop injection and chromatographed at a flow rate of 500 nl/min using a linear gradient of 0–60% of solvent B over solvent A for 40 min. Throughout the separation, ionized molecules were scanned in a positive polarity mode over an m/z range of 108–400. Collision-induced dissociation of each 2′-O-methylnucleoside was carried out with a collision energy +25 and scanned in an m/z range of 50–300.
Circular dichroism spectroscopy and melting experiments:
The piRNA precursor (5′GGUGGAGCCCUGGGUAGCUUGCUGGGGUCAAAGGUCCCUUUGUGGAGGGCCAGAGGGCAGCCUAGGGCCUCUACU3′) or DNA duplexes (5′GTGCTTGGAGCTCATTGGTCCTTGTGTG3′; 5′CACACAAGGACCAATGAGCTCCAAGCAC3′) were synthesized and diluted in 10 mm phosphate buffer (pH 7.4) to a concentration of ∼5 μm. Circular dichroism (CD) spectra were obtained with a JASCO J800 spectropolarimeter (JASCO, Easton, MD). CD melting profiles were recorded at 260 nm while temperature was increased at a rate of 1°/min.
RESULTS
Quantitative profile of differentially expressed piRNAs in testis and ovary:
Although differences between male and female piRNAs have been described before (Houwing et al. 2007), comparative studies of genomic origin, organization, and expression levels between testis and ovary have not previously been performed. We made a piRNA library from zebrafish ovary and produced two sets of microfluidic chips, each consisting of 3466 piRNAs selected randomly from our piRNA pool (http://dev.whu.edu.cn/database/piRNA/, password: 87889294). piRNAs from testis or ovary were labeled and hybridized onto the microarray chips, respectively, in reciprocal cross-labeling experiments. The microarray analysis showed that differential expression signals of piRNAs were apparent between testis and ovary (Figure 1).
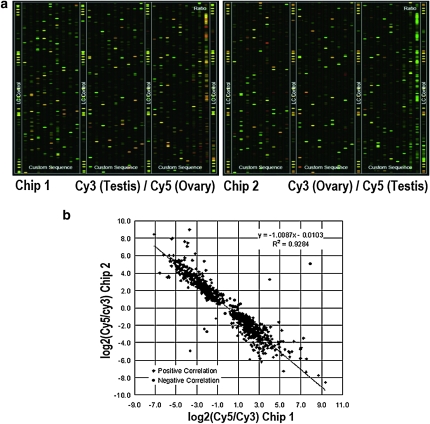
Figure 1.—
(a) Representative regions of piRNA chip images. piRNAs from testis or ovary were labeled with Cy3 or Cy5, respectively, and used as probes to hybridize onto the testis or ovary piRNA chips. The images are displayed in pseudo colors so as to expand the visual dynamic range. As signal intensity increases from 1 to 65,535, the corresponding color changes from blue to green, then to yellow and to red. When the Cy3 level is higher than the Cy5 level, the color is green; when the Cy3 level is equal to the Cy5 level, the color is yellow; and when the Cy5 level is higher than the Cy3 level, the color is red. (b) Signal correlation analysis in two sets of chips. A strong correlation (R2 = 0.9284) between log2 (Cy3/Cyt5) in the two chips was observed.
Fluorescent quantitative analysis of the expression of each piRNA further showed a quantitative difference from 1.58 to 64019.26, exhibiting little association with piRNA copy numbers (genome hits) (Figure 2). Although most of the piRNAs were expressed without difference in testis and ovary, a subset of piRNAs, >20%, did exhibit obvious differential expression between testis and ovary, which clearly fell into two classes: either higher expression in testis or higher expression in ovary (Figure 3).
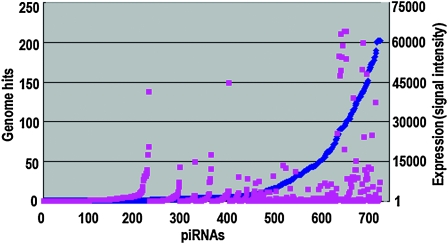
Figure 2.—
Hit numbers (copy numbers) in the genome and expression level of the sex-differentially expressed piRNAs (739). Hit numbers of piRNAs ranged from 1 to 202 (blue dots, y-axis on the left). The expression level of each piRNA is shown in signal intensity (1.58–64,019.26) on the right (red dots, y-axis). The x-axis indicates piRNAs.
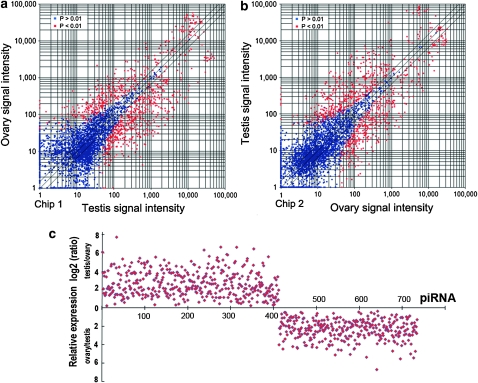
Figure 3.—
Differential expression of piRNAs in testis or ovary in fluorescent intensity. Total piRNAs from testis or ovary were labeled by Cy3 or Cy5, respectively, and used as probes to hybridize onto two independent chips (a and b). Red spots indicate the differentially expressed piRNAs with P-value <0.01 between testis and ovary. Blue spots represent the piRNAs without remarkable difference. (c) Relative expression of differential piRNAs in testis or ovary in signal intensity (log2 value). Relative expression level [testis/ovary (top) or ovary/testis (bottom)] is showed in the y-axis. piRNAs (739) was shown in the x-axis.
Sex-specific piRNAs are derived from different repeat elements:
We next addressed the question of whether or not testis or ovary piRNAs are transcribed from different genomic loci. By mapping these sex-differential piRNAs from chip analysis to the zebrafish genome using the Blat search tool, we found that the piRNAs that exhibited either testis or ovary high expression were derived from different loci. The piRNAs of high expression in testis are mainly derived from LTR1-DR, TDR2, SINE3-1, CR1-4-DR, Dr001038, and Dr000502; the piRNAs of higher expression in ovary were mapped to Ngaro1-DR, EnSpmN1-DR, L1-3-DR, Dr001087, and Dr000417, and a majority of these piRNAs mapped to repeat elements (Table 1).
TABLE 1.
piRNA loci in the genome
| High expression in ovary | High expression in testis | |
|---|---|---|
| Genome hits | 673 (60) | 3446 (71) |
| Repeat hits | 502 | 2391 |
| LTR | 105 | 1386 |
| Ngaro1_DR | 86 | 14 |
| LTR1_DR | 0 | 1368 |
| DNA | 72 | 205 |
| TDR2 | 0 | 202 |
| EnSpmN1_DR | 57 | 0 |
| SINE | 0 | 125 |
| SINE3-1 | 0 | 125 |
| LINE | 11 | 41 |
| L1-3_DR | 9 | 1 |
| CR1-4_DR | 0 | 40 |
| Unknown | 308 | 633 |
| Dr001087 | 103 | 0 |
| Dr000417 | 118 | 0 |
| Dr001038 | 0 | 163 |
| Dr000502 | 0 | 141 |
The repeat-associated piRNAs have been split into five categories: those matching LTR, DNA transposons, SINE, LINE, and unknown repeats. piRNA numbers of each group are indicated in parentheses. Categories and abundance of piRNAs differed between testis and ovary, and the testis-specific loci are boldface type. These piRNAs are cloned in this study and from piRNA chip analysis, with remarkable expression difference between testis and ovary.
Ovary piRNAs tend to form groups:
We next investigated whether or not piRNAs are discrete from one another or fall into groups related to origin. On the basis of sequence homology of piRNAs, we found that piRNAs (the piRNA data are from both this study and Houwing et al. 2007) can be divided into a large number of groups, in which piRNAs showed >95% sequence identity in their 5′ regions (first 70% nucleotides of each piRNA sequence). Table 2 shows the first 20 groups, each with 18 to 92 members. Group 1 has the most at 92 piRNAs, and all are ovary specific. Alignment of piRNA sequences and their flanking genomic sequences of the 92 members showed high sequence similarity among these piRNAs, especially in their 5′ end sequences. More than 33% of ovary piRNAs could be divided into groups, but only 20% of testis piRNAs fell into groups. In addition, all but 1 of the 20 groups were exclusively or dominantly expressed in ovary, especially groups 1, 4, 6, 16, and 19, while only group 10 was dominantly expressed in testis (Table 2). This finding suggests that ovary piRNAs tend to originate from piRNA groups with many members, implying a sex-specific arrangement and organization of piRNA genes in the zebrafish genome.
TABLE 2.
Sex-specific groups of piRNAs
| Testis |
Ovary |
Testis and ovary |
|||||
|---|---|---|---|---|---|---|---|
| Group | Total no. | No. | % | No. | % | No. | % |
| 1 | 92 | 0 | 0.00 | 92 | 100.0 | 0 | 0.00 |
| 2 | 59 | 2 | 3.39 | 54 | 91.53 | 3 | 5.08 |
| 3 | 35 | 3 | 8.57 | 30 | 85.71 | 2 | 5.71 |
| 4 | 34 | 0 | 0.00 | 34 | 100.0 | 0 | 0.00 |
| 5 | 34 | 4 | 11.76 | 30 | 88.24 | 0 | 0.00 |
| 6 | 33 | 0 | 0.00 | 33 | 100.0 | 0 | 0.00 |
| 7 | 27 | 0 | 0.00 | 25 | 92.59 | 2 | 7.41 |
| 8 | 27 | 10 | 37.04 | 5 | 18.52 | 12 | 44.44 |
| 9 | 26 | 2 | 7.69 | 21 | 80.77 | 3 | 11.54 |
| 10 | 25 | 19 | 76.00 | 1 | 4.00 | 5 | 20.00 |
| 11 | 24 | 1 | 4.17 | 21 | 87.50 | 2 | 8.33 |
| 12 | 24 | 2 | 8.33 | 22 | 91.67 | 0 | 0.00 |
| 13 | 23 | 1 | 4.35 | 20 | 86.96 | 2 | 8.70 |
| 14 | 22 | 2 | 9.09 | 16 | 72.73 | 4 | 18.18 |
| 15 | 21 | 1 | 4.76 | 20 | 95.24 | 0 | 0.00 |
| 16 | 20 | 0 | 0.00 | 20 | 100.0 | 0 | 0.00 |
| 17 | 20 | 1 | 5.00 | 18 | 90.00 | 1 | 5.00 |
| 18 | 19 | 4 | 21.05 | 13 | 68.42 | 2 | 10.53 |
| 19 | 19 | 0 | 0.00 | 19 | 100.0 | 0 | 0.00 |
| 20 | 18 | 3 | 16.67 | 12 | 66.67 | 3 | 16.67 |
The piRNAs are divided into groups, the first 20 of which are shown here. piRNAs in groups 1–7, 9, 11–13, 15–17, and 19 were exclusively or dominantly expressed in ovary, while only group 10 piRNAs were dominantly expressed in testis. Ovary-specific groups (1, 4, 6, 16, and 19) are in boldface type. The piRNA data are from both this study and Houwing et al. (2007).
Secondary piRNA structures in the 3′ end:
To explore whether the piRNAs show any evidence of secondary structure, we used the genomic location information (11,000 loci) of piRNAs with sex-differential expression from our chip analysis, to extract 400-nt sequences that are from 200 nt upstream to 200 nt downstream of the piRNAs. Mfold (version 3.2) software was used to predict the secondary structure of the extracted regions. As compared to a general frequency of 0.55 paired nucleotides in the genome, we observed an obvious folding region around the 3′ end of piRNA sequence (pair frequency of up to 0.7) (Figure 4b), which is nearly comparable to the typical folding of zebrafish miRNAs (Figure 4a). The 3′ end of the piRNA sequences may pair with nearby 3′ sequences or with downstream or upstream genomic sequences. The 3′ ends of piRNA are usually involved in pairing, so a high proportion of folding occurs in the 3′ end of the piRNAs (Figure 4, b and d).
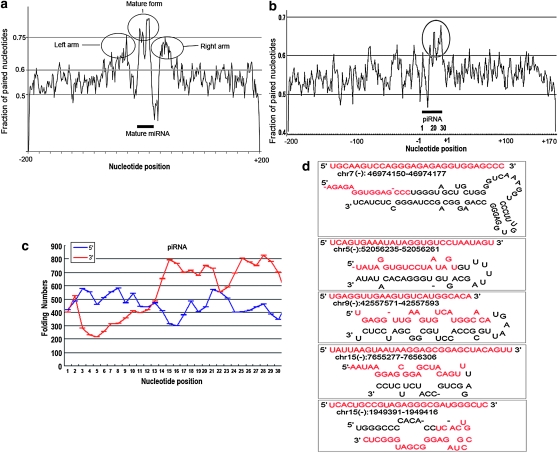
Figure 4.—
Folding analysis of piRNAs and miRNAs. The fraction of paired bases of each position along the 400-nt genomic sequences of all miRNAs (266) and piRNAs (739), which was calculated using the folding structure with minimum free energy, was shown in (a) and (b) respectively. Secondary structure of 400-nt sequences around miRNAs and piRNA in both upstream and downstream genomic locations using Mfold (version 3.2) software. The y axis indicates the fraction of paired nucleotides. Upstream nucleotides of piRNA are shown from +1 to +200, and downstream from −1 to −170. The piRNA and miRNA regions are indicated by the black bar, and the folding region is circled. Folding regions were circled. (c), Folding analysis of the 6-nt paired sequences by comparing the piRNA sequence with 200 nt upstream (blue line) or downstream (red line) flanking sequences. (d) Five examples of piRNA secondary structures. Genomic location formation was indicated below each piRNA sequence. Red letters showed the piRNA sequences.
To further see whether piRNA folds dominantly with up- or downstream genomic sequence, we calculated the 6-nt paired sequence (4–6 bp were defined as a paired sequence within the 6-nt sequence) numbers at each position along the piRNA sequence by scanning and comparison with the up- or downstream 200-nt sequences based on the folding structure with minimum free energy. A markedly high fraction of pairing from around nucleotide 13 of piRNAs with their downstream genomic sequences was observed; however, the same analysis did not show remarkably pairing in their upstream genomic sequences (Figure 4c), further indicating that the piRNA pairing mostly occurs between the 3′ end and its downstream genomic sequence. Five examples of folding structures of the piRNA precursors are listed in Figure 4d.
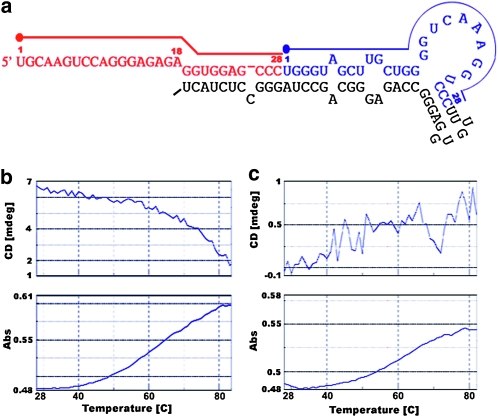
To further confirm formation of folding structures, we synthesized a piRNA precursor (Figure 5a) and tested both the CD spectral and melting curves of the RNA, which revealed a typical folding curve and confirmed that the piRNA precursors form a secondary structure in vitro (Figure 5, b and c).
Figure 5.—
Typical folding of ovary-specific piRNA precursor from chromosome 19 (a) and CD spectral and absorbance (melting curve) of pre-piRNA with secondary structure (b) or DNA duplex as a control (c) at 260 nm in a temperature range from 28 to 84°C. Different colors represent various piRNAs derived from the predicted precursor. Dots indicate the start sites of piRNAs.
Detection of sex-specific piRNAs:
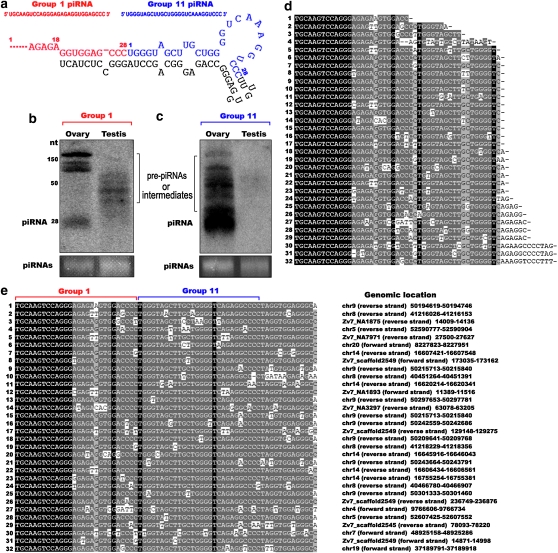
To further explore sex specification of piRNA expression, we analyzed the ovary-specific piRNAs from group 1 using Northern hybridization and noncoding RNA cloning. When using Northern hybridization with the piRNA from group 1 as a probe (Figure 6a), we detected mature piRNAs only in ovary; however, immature intermediates or precursors, but not mature piRNAs, were observed in testis (Figure 6b). In addition, using the 3′ flanking region from group 1 as a probe, we only detected both mature piRNAs and immature intermediates in ovary (Figure 6c). Even though the probe may hybridize with the other piRNAs of the group as probe homology, these piRNAs from the longer bands are also immature, as they are out of the piRNA size. Interestingly, the 3′ flanking region of the group 1 precursor also generated mature piRNAs that belonged to another group, group 11. This group has at least 24 members in the zebrafish genome, most of which (87.5%) were ovary specific (Table 2). We further confirmed the occurrence of these pre-piRNAs and intermediates by cloning and sequencing (Figure 6d, and their genomic sequences are shown in Figure 6e), and alignment of the sequences showed diverse 3′ ends in both groups, leading us to speculate that piRNAs are generated by cleavage at the 3′ end. These results suggest sex-specific expression of piRNA genes.
Figure 6.—
Detection of sex-specific expression of the piRNAs. (a) Group 1 pre-piRNA folding with the 3′-flanking sequence. (b and c) Mature piRNAs present only in ovary. Northern blotting with group 1 piRNA as a probe (a, red) or with the 3′-flanking region of group 1 as a probe (a, blue) from both testis and ovary RNAs. The bottom panel shows the loaded piRNAs as a control. Pre-piRNAs, processed intermediates, and piRNAs are indicated. (d) Alignment of sequenced pre-piRNAs and intermediates (1–32) shows that piRNAs from both groups 1 and 11 are processed as a single transcript with diverse 3′ ends in group 1. (e) Alignment of piRNA genomic sequences within members of group 1 (1–32) shows that group 1 piRNAs are conserved, especially in their 5′-end sequences. Their chromosome mapping information is shown on the right.
piRNAs in both testis and ovary of zebrafish are 2′-O-methylated at their 3′ ends:
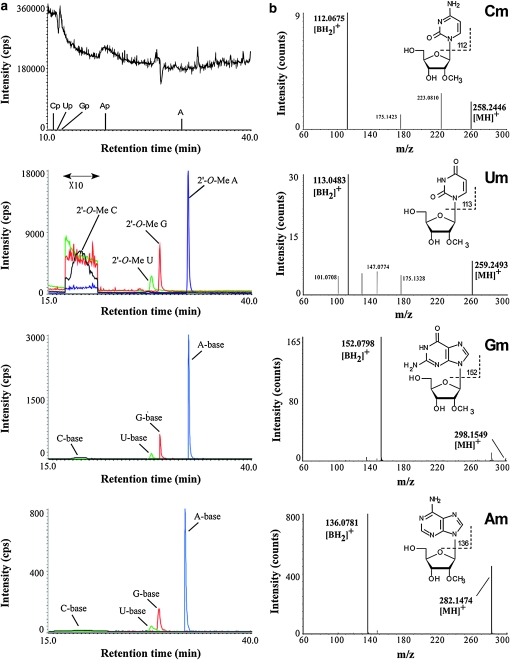
Thus far, piRNA methylation has been tested only in mammalian testis and Drosophila embryo (Horwich et al. 2007; Houwing et al. 2007; Kirino and Mourelatos 2007b; Ohara et al. 2007; Saito et al. 2007). It remains to be confirmed whether zebrafish piRNAs are methylated as in mammals and especially whether they are methylated in the ovary. To identify the terminal modification of piRNAs in zebrafish, total RNAs from testes and ovaries were resolved by denaturing polyacrylamide gel electrophoresis, and piRNA fractions were gel purified and digested using RNase T2, which cleaves after all four residues. In this treatment, most of the residues turned to be nucleoside 3′ phosphate, but only 3′-terminal residues were converted to nucleosides without phosphate groups (Ohara et al. 2007; Suzuki et al. 2007). We analyzed the results of the digestions using capillary liquid chromatography and mass spectrometry as previously described (Ohara et al. 2007; Saito et al. 2007). Four species of 2′-O-methyl nucleosides were clearly observed as major products in piRNAs from both testes and ovaries (Figure 7a). We further analyzed each proton adduct of 2′-O-methyl nucleoside with MS/MS using collision-induced dissociation. Selected reaction monitoring for base-related product ions (Figure 7a) and the mass spectrum (Figure 7b) of each 2′-O-methyl nucleoside also confirmed the presence of a methyl group attached to each ribose portion. To eliminate the possibility that these were 3′-O-methyl nucleosides, RNase T2-digested piRNAs were co-injected with a series of synthetic 3′-O-methyl nucleosides as reported previously (Saito et al. 2007). Each 3′-O-methyl nucleoside was eluted at distinct retention times from each 3′-terminal nucleoside of the piRNAs (data not shown). In addition, each synthetic 2′-O-methyl nucleoside was eluted at the same retention time with its corresponding 3′-terminal nucleoside of the piRNAs (data not shown). Results showed that they are almost entirely methylated at the 2′-O-hydroxyl group on the ribose of the last nucleotide.
Figure 7.—
piRNAs from zebrafish testes and ovaries are 2′-O-methylated at their 3′ ends. (a) First panel, total ion chromatogram of the testis piRNAs digested by RNase T2. The retention times of the observed 3′ phosphonucleotides (Cp, Up, Gp, and Ap) and adenosine (A) are indicated. The considerable amount of adenosine observed is due to contamination, as indicated in the analysis of mouse piRNA (Ohara et al. 2007). Second panel, mass chromatograms for proton adducts of the testis piRNA 3′-terminal nucleosides, 2′-O-Me C (m/z 258, black line), 2′-O-Me U (m/z 259, green line), 2′-O-Me G (m/z 298, red line), and 2′-O-Me A (m/z 282, blue line). Third panel, mass chromatograms of selected reaction monitoring for base-related product ions of 2′-O-Me C (m/z 112, black line), 2′-O-Me U (m/z 113, green line), 2′-O-Me G (m/z 152, red line), and 2′-O-Me A (m/z 136, blue line) of piRNAs from testes. Fourth panel, the same SRM chromatograms as in the third panel for piRNAs from ovaries. (b) CID spectra for 2′-O-Me C (Cm), 2′-O-Me U (Um), 2′-O-Me G (Gm), and 2′-O-Me A (Am). The base-related product ion (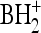 ) of each 2′-O-methylnucleoside is indicated.
) of each 2′-O-methylnucleoside is indicated.
DISCUSSION
Sex-specific genomic organization of the piRNA genes in zebrafish:
Although Dmy/Dmrt1y, a key factor for sex determination, has been identified in medaka (Matsuda et al. 2002; Nanda et al. 2002), the mechanisms of sex determination and differentiation in fish remain largely unclear (Sekido and Lovell-Badge 2009). More factors that contribute to sex determination and differentiation need to be identified. Recently, sex-specific organization of piRNAs has been found in Drosophila (Li et al. 2009; Malone et al. 2009). Our findings that a sex-specific organization of piRNAs also exists in zebrafish suggest that sex-specific organization of piRNAs may be conserved between vertebrate and fly. Furthermore, the expression of piRNA in primary oocytes and developing male germ cells suggests that piRNA may be involved in sexual development (Houwing et al. 2007).
Biogenesis and functions of zebrafish piRNAs:
Aspects of piRNA biogenesis and functions have also been mysterious. On the basis of the features of piRNAs in Drosophila, two teams proposed the ping-pong model for piRNA production (Brennecke et al. 2007; Gunawardane et al. 2007). On the basis of the model, the complementary relationship between sense and antisense piRNA populations suggests an amplification cycle for piRNAs where sense piRNAs in Ago3 cleave long antisense RNA and guide the formation of the 5′ end of antisense piRNAs in Aub (the piRNA-binding protein Aubergine). In the amplification loop, the 5′ ends of piRNAs may be defined by Slicer activity. The most important premise of this model is that the AGO3-associated piRNAs should be complementary to the first 10 nt of Aub- and Piwi-associated piRNAs. Indeed, 20% of all piRNAs have a partner with a 5′ end that can be mapped 10 nucleotides away on the complementary strand in Drosophila (Brennecke et al. 2007). However, recent studies in Drosophila show an Ago3-independent piRNA pathway centered on Piwi in somatic cells, in the absence of ping-pong amplification (Li et al. 2009; Malone et al. 2009).
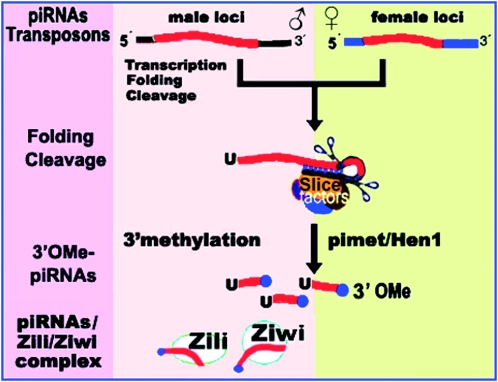
While the ping-pong model may explain piRNA production in zebrafish, other mechanisms of piRNA generation cannot be excluded. Our results show preferential folding at the 3′ end of piRNA precursor, which may favor the cleavages of the piRNA precursor to generate mature piRNAs. After the piRNA processing, methylation modification occurs at the 2′-O-hydroxyl group on the ribose of the last nucleotide, mediated by a common piRNA methyltransferase, Pimet/Hen1 (Figure 8). How does a mechanism keep the sex specificity of piRNA expression? Both the expression and the processing-regulation of piRNA genes may ensure the sex specificity of piRNA genes and their appearance in the specific gonad.
Figure 8.—
The 3′-end processing model of piRNA biogenesis. Pre-piRNA precursors are transcribed from separate genomic loci in males and females, forming a hairpin structure at the 3′ end, followed almost simultaneously by cleavages to produce piRNAs. The piRNAs are modified at the 2′-O-hydroxyl group on the ribose of the last nucleotide, mediated by a common piRNA methyltransferase, Pimet/Hen1. After transcription, folding, cleavage, and methylation, the piRNAs may be amplified through the “ping-pong” mechanism; in addition, there may also be another unknown mechanism for amplification in females. The 3′O-Me-piRNAs exert their role in spermatogenesis and genomic stability by interaction with Zili/Ziwi.
The features of a common 5′ U end and a 3′ end of variable length further support the 3′-processing model of piRNAs; this model may be similar to the tRNA half cleavage that occurs under a variety of stress conditions (Thompson and Parker 2009). The piRNA processing may occur immediately following primary transcription of piRNA genes or transposons in mammals, as it is scarcely detectable for a long precursor piRNA in testis by the RACE analysis (Aravin et al. 2006); however, long piRNA transcripts have also been observed in mouse testis (Ro et al. 2007). Because piRNA biogenesis is independent of Dicer (Houwing et al. 2007), the processing of piRNAs at the 3′ end may be similar to that of the Slicer-mediated process, or may require other proteins or RNAs. Identification of such molecules involved in piRNA processing will help us better understand the functions of piRNA. Although the 3′ processing may be similar to tRNA half cleavage, piRNAs go through additional modifications, such as 3′ methylation followed by the interaction with Zili/Ziwi, before exerting their role in spermatogenesis.
The role of piRNAs remains rather unclear during oogenesis. Nevertheless, ovarian piRNAs preferentially form groups compared to testicular piRNAs, suggesting that piRNAs involving oogenesis have unique, distinct properties compared to those involving spermatogenesis. piRNAs may function in chromosomal remodeling during oocyte meiosis because the dispersed distribution of piRNAs within the whole genome facilitates their interactions with each chromosome (Mochizuki et al. 2002). Alternatively, piRNAs may act as a factor in the nuclear or cytoplasmic skeleton. Earlier results support this view: the attachment of small molecular-weight RNA species (smwRNAs) to the nuclear skeleton (Miller et al. 1978), the localization of Doc RNA (retrotransposon) in the region of the cytoskeleton of the Drosophila oocyte (Zhao and Bownes 1998), and the localization of piRNA in the male mammalian meiotic nucleus (Marcon et al. 2008). Cofactors may also influence piRNA functions; none has yet been identified in the ovary.
Is 2′-O-methylation of the piRNA 3′ terminus a conserved feature?
The question of whether 3′-end methylation occurs in zebrafish piRNAs, especially ovarian piRNAs, is intriguing. Presently, piRNA methylation has only been detected in mammalian testis and Drosophila embryo (Horwich et al. 2007; Kirino and Mourelatos 2007b; Ohara et al. 2007; Saito et al. 2007). The possibility of methylation in zebrafish has been proposed (Houwing et al. 2007). We have shown that piRNAs from both testis and ovary are entirely methylated. This identical methylation of piRNAs has been reported in mouse, rat, and Drosophila, suggesting evolutionary conservation of the 3′ terminal modification pattern of piRNAs, regardless of whether they are derived from ovary or testis. Moreover, the methylation modification seems to be mediated by a common piRNA methyltransferase, Pimet/DmHen1/mHen1 (the homolog of Arabidopsis HEN1 methyltransferase), in Drosophila and mouse (Horwich et al. 2007; Kirino and Mourelatos 2007a; Saito et al. 2007). However, the function of 3′-end methylation of piRNAs is largely unknown, although methylation may stabilize piRNAs (Kurth and Mochizuki 2009). Two lines of evidence appear to be contradictory: Arabidopsis became less fertile due to mutations in hen1 (Chen et al. 2002), whereas Drosophila remained viable and fertile despite mutations in Pimet (Saito et al. 2007). Extensive analyses of putative cofactors and the mechanism of piRNA methylation will enrich our understanding of piRNA biogenesis and function.
Acknowledgments
This work was supported by the National Natural Science Foundation of China, the National Key Basic Research project (2010CB126306), Program of Wuhan Subject Chief Scientist, and the 111 project no. B06018.
References
- Aravin, A., D. Gaidatzis, S. Pfeffer, M. Lagos-Quintana, P. Landgraf et al., 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442 203–207. [DOI] [PubMed] [Google Scholar]
- Aravin, A. A., N. M. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozovsky et al., 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11 1017–1027. [DOI] [PubMed] [Google Scholar]
- Azuma-Mukai, A., H. Oguri, T. Mituyama, Z. R. Qian, K. Asai et al., 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. USA 105 7964–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., A. A. Aravin, A. Stark, M. Dus, M. Kellis et al., 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brodersen, P., and O. Voinnet, 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Carthew, R. W., 2006. Molecular biology. A new RNA dimension to genome control. Science 313 305–306. [DOI] [PubMed] [Google Scholar]
- Chambeyron, S., A. Popkova, G. Payen-Groschene, C. Brun, D. Laouini et al., 2008. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc. Natl. Acad. Sci. USA 105 14964–14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., J. Liu, Y. Cheng and D. Jia, 2002. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, A., R. Sachidanandam, G. J. Hannon and M. A. Carmell, 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442 199–202. [DOI] [PubMed] [Google Scholar]
- Grivna, S. T., E. Beyret, Z. Wang and H. Lin, 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, L. S., K. Saito, K. M. Nishida, K. Miyoshi, Y. Kawamura et al., 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315 1587–1590. [DOI] [PubMed] [Google Scholar]
- Horwich, M. D., C. Li, C. Matranga, V. Vagin, G. Farley et al., 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17 1265–1272. [DOI] [PubMed] [Google Scholar]
- Houwing, S., L. M. Kamminga, E. Berezikov, D. Cronembold, A. Girard et al., 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129 69–82. [DOI] [PubMed] [Google Scholar]
- Houwing, S., E. Berezikov and R. F. Ketting, 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27 2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi, Y., N. Shigi, J. Kato, A. Nishimura and T. Suzuki, 2006. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 21 97–108. [DOI] [PubMed] [Google Scholar]
- Kim, J. K., H. W. Gabel, R. S. Kamath, M. Tewari, A. Pasquinelli et al., 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308 1164–1167. [DOI] [PubMed] [Google Scholar]
- Kim, V. N., J. Han and M. C. Siomi, 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10 126–139. [DOI] [PubMed] [Google Scholar]
- Kirino, Y., and Z. Mourelatos, 2007. a The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13 1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino, Y., and Z. Mourelatos, 2007. b Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 14 347–348. [DOI] [PubMed] [Google Scholar]
- Klattenhoff, C., and W. Theurkauf, 2008. Biogenesis and germline functions of piRNAs. Development 135 3–9. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa, S., T. Watanabe, K. Gotoh, Y. Totoki, A. Toyoda et al., 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth, H. M., and K. Mochizuki, 2009. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, N. C., L. P. Lim, E. G. Weinstein and D. P. Bartel, 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Lau, N. C., A. G. Seto, J. Kim, S. Kuramochi-Miyagawa, T. Nakano et al., 2006. Characterization of the piRNA complex from rat testes. Science 313 363–367. [DOI] [PubMed] [Google Scholar]
- Lee, S. R., and K. Collins, 2006. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 20 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., V. V. Vagin, S. Lee, J. Xu, S. Ma et al., 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, A. K., L. Tao and T. Kai, 2009. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 186 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, C. D., J. Brennecke, M. Dus, A. Stark, W. R. McCombie et al., 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon, E., T. Babak, G. Chua, T. Hughes and P. B. Moens, 2008. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 16 243–260. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417 559–563. [DOI] [PubMed] [Google Scholar]
- Miller, T. E., C. Y. Huang and A. O. Pogo, 1978. Rat liver nuclear skeleton and small molecular weight RNA species. J. Cell Biol. 76 692–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, K., N. A. Fine, T. Fujisawa and M. A. Gorovsky, 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110 689–699. [DOI] [PubMed] [Google Scholar]
- Nakayashiki, H., 2005. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 579 5950–5957. [DOI] [PubMed] [Google Scholar]
- Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al., 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara, T., Y. Sakaguchi, T. Suzuki, H. Ueda and K. Miyauchi, 2007. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 14 349–350. [DOI] [PubMed] [Google Scholar]
- Ro, S., C. Park, R. Song, D. Nguyen, J. Jin et al., 2007. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA 13 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Y. Sakaguchi, T. Suzuki, H. Siomi and M. C. Siomi, 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 21 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido, R., and R. Lovell-Badge, 2009. Sex determination and SRY: Down to a wink and a nudge? Trends Genet. 25 19–29. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Y. Sakaguchi and T. Suzuki, 2007. Mass spectrometric analysis of 3′-terminal nucleosides in non-coding RNAs. Nat. Protoc. doi: 10.1038/nprot.2007.185. [DOI]
- Thompson, D. M., and R. Parker, 2009. Stressing out over tRNA cleavage. Cell 138 215–219. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya, Y., Y. Hao, E. Beyret, H. Yin, S. Kuramochi-Miyagawa et al., 2009. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 284 6507–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, V. V., A. Sigova, C. Li, H. Seitz, V. Gvozdev et al., 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 320–324. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20 759–771. [DOI] [PubMed] [Google Scholar]
- Watanabe, T., A. Takeda, T. Tsukiyama, K. Mise, T. Okuno et al., 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, M. L., Y. Bennasser, K. Watashi, S. Y. Le, L. Houzet et al., 2009. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37 6575–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D., and M. Bownes, 1998. The RNA product of the Doc retrotransposon is localized on the Drosophila oocyte cytoskeleton. Mol. Gen. Genet. 257 497–504. [DOI] [PubMed] [Google Scholar]
- Zhou, R., H. Cheng and T. Tiersch, 2001. Differential genome duplication and fish diversity. Rev. Fish Biol. Fisher. 11 331–337. [Google Scholar]