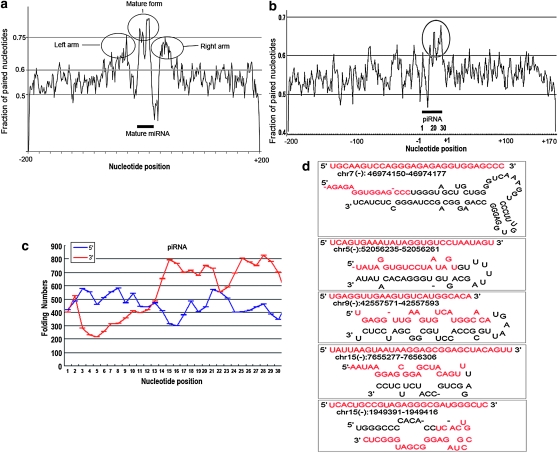
Figure 4.—
Folding analysis of piRNAs and miRNAs. The fraction of paired bases of each position along the 400-nt genomic sequences of all miRNAs (266) and piRNAs (739), which was calculated using the folding structure with minimum free energy, was shown in (a) and (b) respectively. Secondary structure of 400-nt sequences around miRNAs and piRNA in both upstream and downstream genomic locations using Mfold (version 3.2) software. The y axis indicates the fraction of paired nucleotides. Upstream nucleotides of piRNA are shown from +1 to +200, and downstream from −1 to −170. The piRNA and miRNA regions are indicated by the black bar, and the folding region is circled. Folding regions were circled. (c), Folding analysis of the 6-nt paired sequences by comparing the piRNA sequence with 200 nt upstream (blue line) or downstream (red line) flanking sequences. (d) Five examples of piRNA secondary structures. Genomic location formation was indicated below each piRNA sequence. Red letters showed the piRNA sequences.