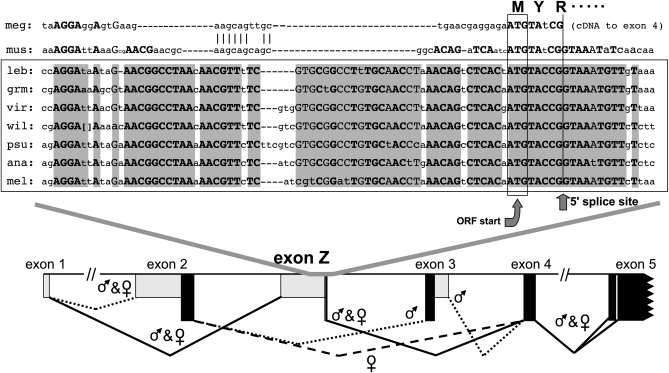
Figure 6.—
DNA sequence conservation near the 5′-splice site of the newly discovered Sxl exon Z. Uppercase letters in boldface type indicate positions completely conserved in a diverse collection of six Drosophila and one Scaptodrosophila species [namely, Scaptodrosophila lebanonensis (leb) and the Drosophila species grimshawi (grm), virilis (vir), wilistoni (wil), pseudoobscura (psu), ananassae (ana), and melanogaster (mel)] (Drosophila 12 Genomes Consortium 2007; Siera and Cline 2008). Uppercase letters in regular type signify positions at which all but one of the seven sequences are identical. For D. wilistoni, the sequence CTCTCTGTAAAGAG was present between the brackets. The boxed ATG is the only potential translation start site for each species that would encode nearly full-length Sxl proteins from mRNAs containing exon Z. A corresponding region was apparent in a cDNA from the scuttle fly, Megaselia scalaris (meg) (Sievert et al. 2000) and in genomic DNA from the housefly, Musca domestica (mus) (our sequence). The splice site shown for Megaselia exon Z is based on the authors' revised genomic sequence (AF110846.1) near exon 4 (only cDNA sequence was available near exon Z). The schematic in the bottom section is for D. melanogaster and shows the sex in which the splice sites for each of the indicated Sxl exons are active. Solid bars indicate protein-coding regions. Note that exon Z mRNAs are made by skipping the male-specific exon 3 in both sexes (see Figure 7).