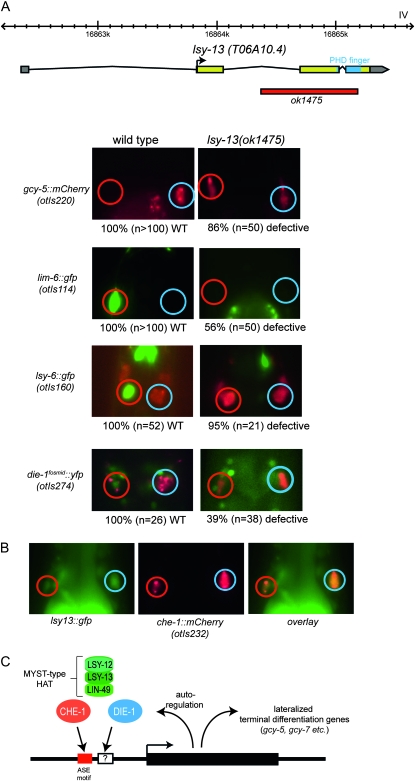
Figure 3.—
The MYST-complex component lsy-13 affects ASE laterality. (A) lsy-13 controls ASE laterality. The upper panel shows the structure of the lsy-13 locus and the lsy-13 null allele. The gene structure is confirmed by EST clones (www.wormbase.org). All other panels show the head regions of adult animals. Numbers below the panels indicate the penetrance of the phenotype, i.e. the fraction of animals that display the phenotype shown in the fluorescent image above. The die-1 and lsy-6 wild-type control images are the same as in other figures and shown for comparison only. The red fluorescent marker in the lsy-6 panel (ceh-36prom∷dsRed2) allows identification of the ASE neurons. Animals that express the die-1 reporter fosmid also contain a ASEL/R-expressed red fluorescent reporter (che-1∷mCherry). A list of transgenes used in the study is provided in File S1. We note that lsy-13 function can be maternally supplied (homozygous offspring of a heterozygous lsy-13 parent does not display a mutant phenotype). In contrast to the lin-49 null mutant animals, lsy-13(ok1475) null mutant animals are viable and display no obvious morphological abnormalities. (B) A reporter gene which contains 2.8 kb of 5′ sequences to the first exon of lsy-13, generated by PCR fusion (Hobert 2002), is broadly expressed, including in the two ASE neurons, marked with a red fluorescent reporter gene. Primer sequences for the construct are provided in File S1. (C) Model for lsy-12/lsy-13/lin-49 function, based on the phenotypic similarities between the genes shown here. che-1 directly regulates expression of terminal differentiation genes—both symmetrically and asymmetrically expressed ones—as well as regulators of the bistable feedback loop (Etchberger et al. 2007, 2009). Left/right asymmetrically expressed genes, contain cis-regulatory elements (indicated by “?”) in addition to the CHE-1 binding site (the ASE motif) that restrict CHE-1 activity to ASEL or ASER (Etchberger et al. 2009). Genetically, die-1 controls the activity of the factors that restrict che-1 activity, but it is not known whether die-1 fulfills this function directly (through binding to these additional motifs) or indirectly through the regulation of other factors. Since die-1 autoregulates its own transcription (L. Cochella and O. Hobert, unpublished data), the HAT complex also impinges on die-1 expression itself.