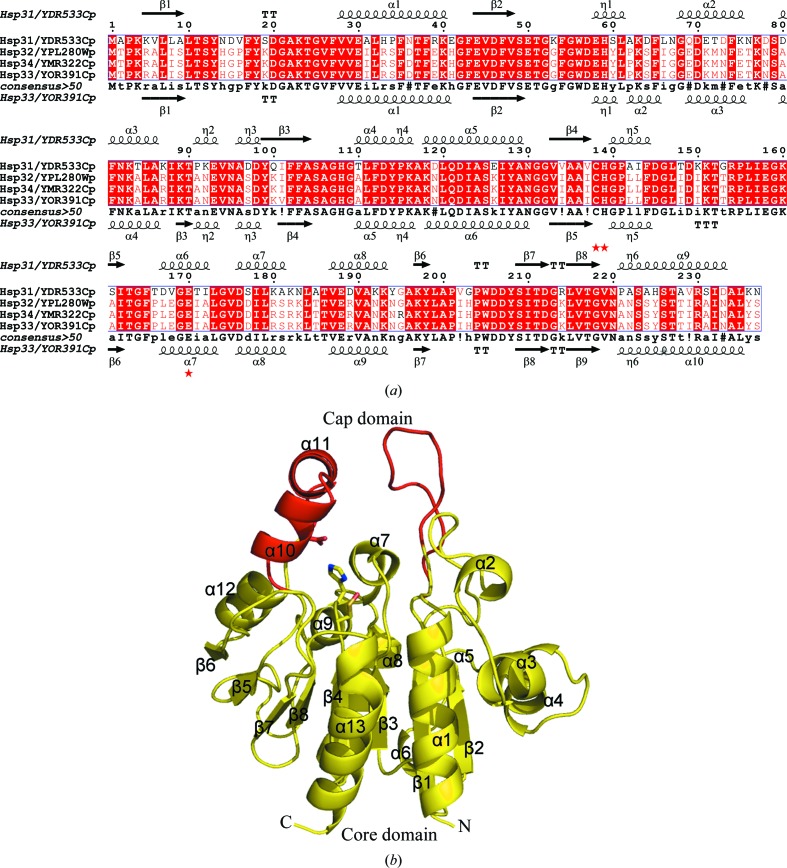
Figure 1.
Sequence alignment and overall structure. (a) Structure-based sequence alignment of Hsp33, Hsp31, Hsp32 and Hsp34 from S. cerevisiae. The secondary structure of Hsp31 (PDB codes 1qvz and 1rw7; Wilson et al., 2004 ▶; Graille et al., 2004 ▶) is shown at the top and that of Hsp33 (PDB code 3mii) is shown at the bottom. The residues involved in the catalytic triad are marked by red stars. Alignments were performed using the programs MULTALIN (Corpet, 1988 ▶) and ESPript (Gouet et al., 2003 ▶). (b) Cartoon representation of the yeast Hsp33 monomer. The core and cap domains are coloured yellow and red, respectively. The residues constituting the putative catalytic triad are shown as sticks. All figures were prepared using PyMOL (DeLano, 2002 ▶).