Table 1. Data-collection statistics for the crystal of yeast Hsp33.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P43212 |
| Unit-cell parameters (, ) | a = b = 94.4, c = 132.2, = = = 90 |
| Molecules per asymmetric unit | 2 |
| Resolution range () | 60.632.40 (2.532.40) |
| Unique reflections | 45873 (6819) |
| Completeness (%) | 97.3 (99.4) |
| I/(I) | 8.7 (2.3) |
| R merge † (%) | 11.8 (44.3) |
| Average redundancy | 2.3 (2.2) |
| Structure refinement | |
| Resolution range () | 25.502.40 (2.462.40) |
| R factor‡/R free § | 0.222/0.246 |
| No. of protein atoms | 3635 |
| No. of heteroatoms | 31 |
| No. of water atoms | 168 |
| R.m.s.d.¶ bond lengths () | 0.006 |
| R.m.s.d. bond angles () | 0.946 |
| Mean B factor (2) | 21.74 |
| Ramachandran plot†† | |
| Most favoured (%) | 97.19 |
| Additional allowed (%) | 2.81 |
| Outliers (%) | 0 |
| Poor rotamers (%) | 0.27 |
| Clash score | 5.9 |
| PDB code | 3mii |
R
merge = 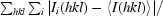
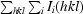 , where I
i(hkl) is the intensity of an observation and I(hkl) is the mean value for the unique reflection; summations are over all reflections.
, where I
i(hkl) is the intensity of an observation and I(hkl) is the mean value for the unique reflection; summations are over all reflections.
R factor = 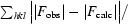
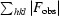 , where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively.
, where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively.
R free was calculated with 5% of the data excluded from refinement.
Root-mean-square deviation from ideal values (Engh Huber, 1991 ▶).
Categories as defined by MolProbity (Chen et al., 2010 ▶).
