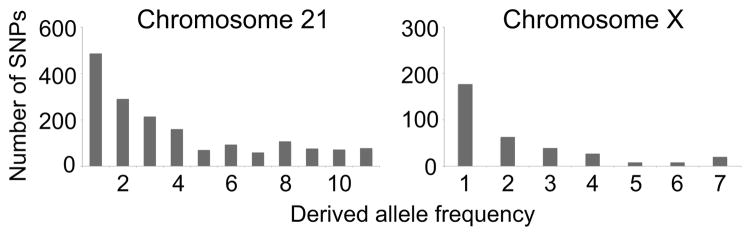
Fig. 2. Derived allele frequency distributions.
For all segregating sites with filtered read coverage sufficient for SNP identification in the results from the fecal DNA samples of all individuals, we determined how many chromosomes (n = 12 total for chromosome 21; n = 8 total for chromosome X) carried the derived allele (derived allele frequency). Plots depict the number of SNPs in each derived allele frequency bin. Ancestral/derived allele states were estimated by comparisons to the human and rhesus macaque reference genome sequences.