Abstract
Understanding the secondary structure of peptides is important in protein folding, enzyme function, and peptide-based drug design. Previous studies of synthetic Ala-based peptides (>12 a.a.) have demonstrated the role for charged side chain interactions involving Glu/Lys or Glu/Arg spaced three (i, i + 3) or four (i, i + 4) residues apart. The secondary structure of short peptides (<9 a.a.), however, has not been investigated. In this study, the effect of repetitive Glu/Lys or Glu/Arg side chain interactions, giving rise to E-R/K helices, on the helicity of short peptides was examined using circular dichroism. Short E-R/K–based peptides show significant helix content. Peptides containing one or more E-R interactions display greater helicity than those with similar E-K interactions. Significant helicity is achieved in Arg-based E-R/K peptides eight, six, and five amino acids long. In these short peptides, each additional i + 3 and i + 4 salt bridge has substantial contribution to fractional helix content. The E-R/K peptides exhibit a strongly linear melt curve indicative of noncooperative folding. The significant helicity of these short peptides with predictable dependence on number, position, and type of side chain interactions makes them an important consideration in peptide design.
Keywords: α-helix, E-R/K peptides, peptide design, salt bridges, circular dichroism
Introduction
Understanding and modifying secondary structure in peptides is necessary in understanding protein folding,1 in engineering proteins,2 and in designing enzymatic targets.3 Since the discovery of significant helicity in the 13 amino acid C-peptide from ribonuclease A,4,5 there has been a considerable amount of research on helicity of de novo peptides. Peptides ranging from 12 to 30 residues have been studied to understand the interplay between side chain interactions, the helix dipole, and the intrinsic helix-forming ability of each amino acid.6 This research has revealed that Ala-based peptides show substantial helix formation, as Ala has a high helix propensity.7
Ala-based peptides have been used to look at side chain interactions, such as Glu with Lys (E/K) salt bridges which form stabilizing i → i + 3 and i → i + 4 interactions.8 In Ala-based peptides containing i → i + 4 E/K salt bridges (EAAAK)n, replacing Lys with Arg was found to enhance the helical content of the peptides.9 Studies were also done to examine peptides (≥16 amino acids) that have repetitive Glu-Lys and/or Glu-Arg interactions (which we refer to as the E-R/K motif). For example, Lyu et al.10 examine one 18-residue peptide consisting of a repeating motif of four negatively charged Glu residues followed by four positively charged Lys residues. This motif of four Glu residues followed by a combination of four Lys and/or Arg residues has been found in a variety of proteins, including caldesmon,11 myosin X, and myosin VI.12,13 It has been shown to form long (up to 30 nm), single, stable, and relatively rigid helices (persistence length = 15 nm) in various proteins.14–16
Experimental measurement of helicity using circular dichroism of peptides has been used in conjunction with statistical mechanics models of helix–coil transition17,18 to develop prediction programs for the helicity of peptides. The original Zimm and Bragg19 and Lifson and Roig20 models described the helix–coil transition as a two-step process with helix nucleation followed by helix propagation or elongation. These theories were modified in subsequent studies to include side chain–side chain interactions10 and peptide-capping effects.21 Currently, one of the models, AGADIR, has been benchmarked against the largest selection of peptide sequences.22 AGADIR is an algorithm that is based on helix–coil theory but modified to incorporate experimentally derived parameters.18 The algorithm attempts to obtain an energetic description of the system by splitting the conformational energy of the peptide into a sum of energies: intrinsic helical tendencies of each residue, main chain–main chain hydrogen bonding, side chain–side chain interactions, helical dipole effects, and effects of nonhelical residues.22
While AGADIR predicts the helicity for peptides greater than one helical turn or four residues, there is a lack of experimental data for the helicity of short peptides (<9 amino acids). Ala-based peptides demonstrate only marginal helicity when 10 residues in length.9 As a result of the focus of all helix prediction algorithms on Ala-based peptides, short peptides have largely been overlooked both in experimental and theoretical treatments.
This study shows that peptides shorter than 10 residues can in fact have significant helicity, even without helix-inducing solvents like trifluoroethanol. Short E-R/K–based peptides were studied using circular dichroism. These short peptides exhibited high helicity, and E-R–based E-R/K peptides showed significantly greater helicity than their E-K counterparts.
Results and Discussion
Short peptides demonstrate significant helicity
Peptides with four Glu residues followed by four Lys residues [the (E4K4)n motif] were studied, where n = 1, 2, or 3 [Fig. 1(a)]. All peptides in this study were designed with an N-terminal acetyl group and C-terminal amide cap. A Tyr was placed at the N-terminus separated by a Ser from the rest of the peptide to facilitate concentration measurements.23 Although the peptide bonds contributed by the Tyr and Ser residues are included in all calculations, we will refer to a peptide's length by the number of helix-promoting residues (i.e., excluding Tyr and Ser). Not only were the 16-residue (E4K4)2 and 24-residue (E4K4)3 peptides helical as previously reported10 but also the 8-residue E4K4 peptide demonstrated significant helix content (59%). For the reference helix content of short peptides in the absence of salt bridges, Ala-based peptides were designed with either Arg or Lys for solubility purposes (A1–A3, Table I).
Figure 1.
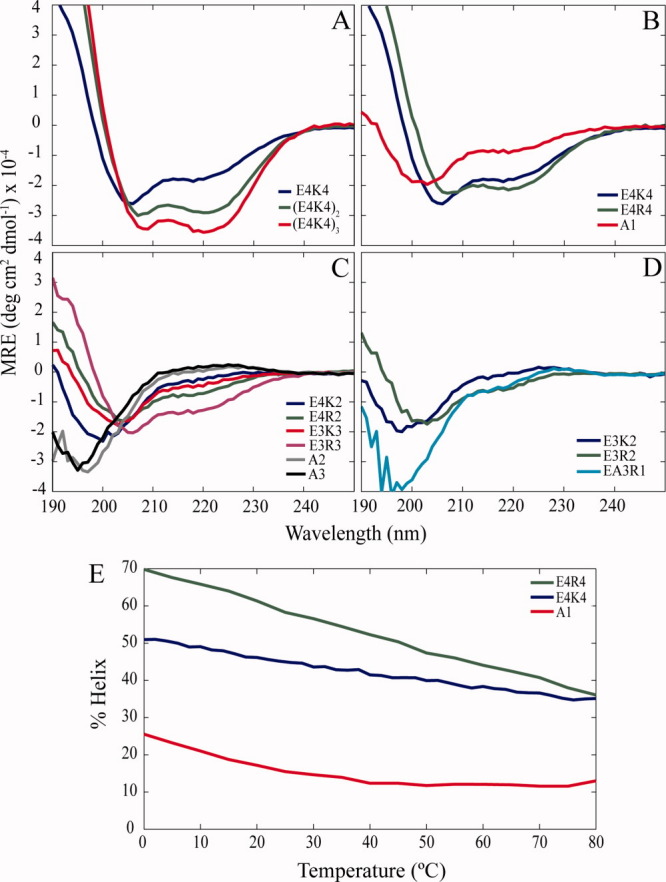
Arg versus Lys in E-R/K peptides. Circular dichroism spectra of (a) EK peptides 8, 16, and 24 residues long, and short E-R/K peptides (b) 8, (c) 6, and (d) 5 residues in length. The isodichroic point at 202 nm and the minima at 208 and 222 nm are characteristic of α-helices. (e) Helix content as a function of temperature (°C) at 222 nm from circular dichroism.
Table I.
Helix Content for Short E-R/K Peptides
| Name | Sequence | [θ]222a | % Helix | AGADIR (%) |
|---|---|---|---|---|
| E4R4 | YSEEEERRRR | −20,100 | 71.2 | 76.6 |
| E4K4 | YSEEEEKKKK | −16,700 | 59.2 | 52.1 |
| E4R2 | YSEEEERR | −6100 | 24.4 | 32.8 |
| E4K2 | YSEEEEKK | −1900 | 7.6 | 11.5 |
| E3R3 | YSEEERRR | −11,900 | 47.6 | 64.0 |
| E3K3 | YSEEEKKK | −3800 | 14.8 | 36.7 |
| E3R2 | YSEEERR | −4200 | 18.6 | 29.9 |
| E3K2 | YSEEEKK | 200 | −0.9 | 13.5 |
| EA3R1 | YSEAAAR | −2400 | 10.2 | 14.8 |
| EA3K1 | YSEAAAK | 700 | −3.1 | 10.3 |
| A1 | YSAAARAARA | −7600 | 26.9 | 22.9 |
| A2 | YSARAARA | 900 | −3.6 | 6.4 |
| A3 | YSAKAAKA | 2100 | −8.4 | 6.4 |
Helix content was calculated from the mean residue ellipticity (MRE) at 222 nm or [θ]222 using the following equation, % helix = 100 ([θ]222/(−39500(1 − 2.57/n))), where n is the number of total peptide bonds and [θ]222 has been corrected for background.22 Helix content values were compared with those predicted by the algorithm AGADIR.
All three peptides have minima at 208 and 222 nm characteristic of α-helices. The isodichroic point at 202 nm is also consistent with a system that occupies two different states, the structured helical state and the unstructured state.8,24 As the helix content of the peptide increases, the 208 nm minimum, which contains contributions from both the helical and the unstructured state, decreases. The helix content is most easily monitored by examining the mean residue ellipticity (MRE) at the 222 nm minimum. To compare our results with AGADIR predictions, the helix content was calculated in accordance with the Chen equation25 used by AGADIR.22 The 1974 Chen equation for calculating percent helix values was later tested with Ala-based peptides of varying chain lengths and was refined slightly.26 In the case of the E4K4 peptide, the helix content is high at 59%, close to the value predicted by the AGADIR algorithm (Table I).
E-R peptides have higher helix content than E-K peptides
To examine the effect of Arg versus Lys in these E-R/K peptides, the E4K4 peptide was initially compared with the E4R4 peptide [Fig. 1(b)]. Not only is the 222 nm minimum at a lower MRE but also the 208 nm minimum has less contribution from the unstructured conformation. From the 222 nm values, the helix content of the E4R4 peptide was calculated to be ∼71% versus 59% for the E4K4 peptide, again fitting well the prediction from the AGADIR algorithm (Table I). The thermal melts of these eight-residue long peptides were noncooperative and melting occurred over a wide temperature range (80°C), as is a characteristic of E-R/K peptides [Fig. 1(e)]. The melts were reversible (data not shown).
As high helicity was observed in eight-residue peptides, we examined even shorter peptides for helix content. Peptides six and five residues in length were examined [Fig. 1(c,d), respectively]. It must be noted that for peptides the unstructured state is a mixture of different backbone conformations, with contributions from polyproline II.24 The polyproline II spectrum is positive between 220 and 230 nm, which is consistent with positive CD values at 222 nm for a few of our peptides, such as A2 and A3.
The helix content for these six and five-residue peptides, listed in Table I, provide two important insights. First, peptides with this E-R/K motif exhibit surprisingly high helix content with only six or even five residues (∼48% for E3R3 and ∼19% for E3R2). Each additional i → i + 4 and i → i + 3 salt bridge increases the helix content of short peptides. In the case of E4R2 and E3R3, the E3R3 peptide, with an additional i → i + 3 salt bridge, has a helix content nearly twice that of E4R2 (Table I). It is worth noting that this increase in helix content likely results some from losing a potentially destabilizing i → i + 4 electrostatic repulsion between the first and fourth E. According to AGADIR, losing this destabilizing interaction, although, only accounts for part (∼60%) of the increase in helix content.
Second, in all cases, the E-R/K peptides that contain Arg have significantly higher helix contents than the corresponding peptides with Lys. For example, E3R3 has ∼48% helix versus ∼15% for E3K3. This trend has been previously reported in the context of Ala-based peptides.9 Knight et al.12 have hypothesized that E-R/K peptides would exhibit greater stability with E-R interactions relative to E-K interactions. The presence of the guanidinium group may enable Arg to interact simultaneously with the negative Glu residues in both directions.13
Conclusions
Secondary structure of short peptides has largely been overlooked because of the general assumption that they are unstructured in solution. This study reports appreciable secondary structure of short peptides that are based on the E-R/K motif. These short peptides are stabilized by i → i + 4 and i → i + 3 salt bridges, with greater helix content in E-R–based peptides relative to E-K peptides. Further studies are needed to understand why E-R–based peptides have higher helix content. The existing prediction program AGADIR, however, did capture this difference between Arg and Lys and compared reasonably with experimental measurements. Overall, this study highlights the importance of the E-R/K motif in determining the secondary structure of short helical peptides and in de novo design of peptides. In the context of longer protein-derived E-R/K single α-helices,14–16 this study suggests that E-R–enriched helices could be more stable than their E-K counterparts. The structural and functional consequences of the relative abundance of R versus K residues in E-R/K helices remain to be determined.
Materials and Methods
Circular dichroism spectroscopy
Peptides were purchased from GenScript and purified to ≥98%, as determined by mass spectrometry and HPLC (GenScript Corp, Piscataway, NJ). Peptide concentrations were calculated using the ɛ280 = 1490 M−1 cm−1.27 CD spectra were acquired using an Aviv 62DS instrument (Aviv Biomedical, Lakewood NJ) with a 1-mm path-length cell. Measurements were taken every 1 nm at 0°C with a 10 s averaging time and with concentrations ranging from 60 to 110 μM in 10 mM sodium phosphate buffer, pH 7.0. MRE was estimated from the following equation, [θ]222 × MRW/[peptide], where MRW is the average molecular weight per residue, [θ]222 is corrected for background, and the peptide concentration is in milligram per milliliter. Melt data were collected every 5°C with a 30 s averaging time and a 2 min equilibration. % Helix was calculated using the equation described in Table I. For all peptides, the reverse melt demonstrated reversibility.
References
- 1.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hua QX, Nakagawa SH, Jia W, Huang K, Philips NB, Hu SQ, Weiss MA. Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications. J Biol Chem. 2008;283:14703–14716. doi: 10.1074/jbc.M800313200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moellering RE, Cornejo M, Davis TN, Bianco CD, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown JE, Klee WA. Helix-coil transition of the isolated amino terminus of ribonuclease. Biochemistry. 1971;10:470–476. doi: 10.1021/bi00779a019. [DOI] [PubMed] [Google Scholar]
- 5.Bierzynski A, Kim PS, Baldwin RL. A salt bridge stabilizes the helix formed by isolated C-peptide of RNase A. PNAS. 1982;79:2470–2474. doi: 10.1073/pnas.79.8.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scholtz JM, Baldwin RL. The mechanism of α-helix formation by peptides. Annu Rev Biomol Struct. 1992;21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- 7.Marqusee S, Robbins VH, Baldwin RL. Unusually stable helix formation in short alanine-based peptides. PNAS. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marqusee S, Baldwin RL. Helix stabilization by Glu−···Lys+ salt bridges in short peptides of de novo design. PNAS. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merutka G, Shalongo W, Stellwagen E. A model peptide with enhanced helicity. Biochemistry. 1991;30:4245–4248. doi: 10.1021/bi00231a020. [DOI] [PubMed] [Google Scholar]
- 10.Lyu PC, Gans PJ, Kallenbach NR. Energetic contribution of solvent-exposed ion pairs to alpha-helix structure. J Mol Biol. 1992;223:343–350. doi: 10.1016/0022-2836(92)90735-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang CL, Chalovich JM, Graceffa P, Lu RC, Mabuchi K, Stafford WF. A long helix from the central region of smooth muscle caldesmon. J Biol Chem. 1991;266:13958–13963. [PMC free article] [PubMed] [Google Scholar]
- 12.Knight PJ, Thirumurugan K, Xu Y, Wang F, Kalverda AP, Stafford WF, III, Sellers JR, Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;208:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 13.Sivaramakrishnan S, Spink BJ, Sim AYL, Doniach S, Spudich JA. Dynamic charge interactions create surprising rigidity in the ER/K α-helical protein motif. PNAS. 2008;105:13356–13361. doi: 10.1073/pnas.0806256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivaramakrishnan S, Sung J, Ali M, Doniach S, Flyvbjerg H, Spudich JA. Combining single-molecule optical trapping and small-angle x-ray scattering measurements to compute the persistence length of a protein ER/K alpha-helix. Biophys J. 2009;97:2993–2999. doi: 10.1016/j.bpj.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baboolal TG, Sakamoto T, Forgacs E, White HD, Jackson SM, Takagi Y, Farrow RE, Molloy JE, Knight PJ, Sellers JR, Peckham M. The SAH domain extends the functional length of the myosin lever. PNAS. 2009;106:22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Süveges D, Gáspári Z, Tóth G, Nyitray L. Charged single alpha-helix: a versatile protein structural motif. Proteins. 2009;74:905–916. doi: 10.1002/prot.22183. [DOI] [PubMed] [Google Scholar]
- 17.Scholtz JM, Qian H, York EJ, Stewart JM, Baldwin RL. Parameters of helix-coil theory for alanine-based peptides of varying chain lengths in water. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 18.Muñoz V, Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. Nat Struct Biol. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 19.Zimm BH, Bragg JK. Theory of phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526–535. [Google Scholar]
- 20.Lifson S, Roig A. On the theory of helix-coil transition in polypeptides. J Chem Phys. 1960;34:1963–1974. [Google Scholar]
- 21.Doig AJ, Chakrabartty A, Klingler TM, Baldwin RL. Determination of the free energies of n-capping in α-helices by modification of the Lifson-Roig helix-coil theory to include N- and C-capping. Biochemistry. 1994;33:3396–3403. doi: 10.1021/bi00177a033. [DOI] [PubMed] [Google Scholar]
- 22.Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem in α-helices: local motifs, long-range electrostatics, ionic-strength dependence, and prediction of NMR parameters. J Mol Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 23.Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry. 1993;32:5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- 24.Shi Z, Woody RW, Kallenbach NR. Is polyproline II a major backbone conformation in unfolded proteins? Adv Protein Chem. 2002;62:163–240. doi: 10.1016/s0065-3233(02)62008-x. [DOI] [PubMed] [Google Scholar]
- 25.Chen YH, Yang JT, Chau KH. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 26.Luo P, Baldwin RL. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 27.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]


