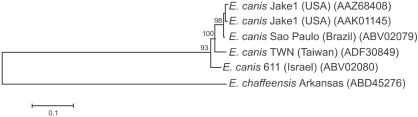
Fig. 4.
Phylogenetic tree based on deduced amino acid sequences of the E. canis gp200. To root the tree, the sequence of an ortholog (ankyrin protein 200) in E. chaffeensis was used as an outgroup. Accession numbers for E. canis isolates and the outgroup species E. chaffeensis are given in parentheses. The lengths of the lines are proportional to the number of amino acid changes. The scale bar at the lower left indicates the number of substitutions per sequence position. The numbers at the nodes represent the percentage of 1,000 bootstrap resamplings.