Abstract
Induced pluripotent stem (iPS) cells are a new alternative for the development of patient-specific stem cells, and the aim of this study was to determine whether differences exist between the cellular and molecular profiles of iPS cells, generated using lentiviral vectors, compared to ES cells. The lentiviral infection efficiency differed according to the method of cell culture (adherent cells: 0.085%; suspended cells: 0.785%). Six iPS cell lines exhibited typical ES cell morphology and marker expression, but varied in their in vitro/in vivo differentiation ability. Global gene transcription analysis revealed that core pluripotency genes were expressed at lower levels in iPS cell lines compared to D3-ES cells (Pou5f1: ×1.6∼2.2-fold, Sox2: ×2.58∼10.0-fold, Eras: ×1.08∼2.54-fold, Dppa5a: ×1.04∼1.41-fold), while other genes showed higher expression in iPS cells (Lin28: ×1.43∼2.33-fold; Dnmt3b: ×1.33∼2.64-fold). This pattern was repeated in a survey of specific functional groups of genes (surface markers, cell death, JAK–STAT and P13K–AKT signaling pathways, endothelial, cardiovascular, and neurogenesis genes). Among the iPS cell lines examined, only two showed similar characteristics to ES cells. These results demonstrated that, in addition to cellular characterization, the numerical evaluation of gene expression using DNA microarrays might help to identify the stem cell stability and pluripotency of iPS cells.
Introduction
Stem cell research is prominent in the fields of biotechnology and medicine as stem cells are recognized as promising donor sources for cell transplantation therapies for diseases such as Parkinson's disease, spinal cord injury, and heart failure (Thomson et al., 1998). Stem cells can be derived either from embryos or from various postnatal sources; the former are known as embryonic stem (ES) cells and the latter as adult stem cells. ES cells are capable of differentiating into cells representing all three germ layers and have prolonged self-renewal capacity, while adult stem cells exhibit limited plasticity and poor growth potential. However, recently developed induced pluripotent stem (iPS) cells, artificially derived from somatic cells reprogrammed by the introduction of certain transcription factors, are paving the way toward simplifying the production of patient-specific stem cells without the controversial use of embryos (Takahashi and Yamanaka, 2006; Yu et al., 2007).
Several researchers have reported that, in many respects, iPS cells are very similar to ES cells, with comparable morphology, surface marker expression, embryoid body formation, epigenetic status, teratoma formation, and direct differentiation into neural cells and beating cardiomyocytes (Maherali et al, 2007; Park et al., 2008; Takahashi et al., 2007). The reprogramming method was initially attempted with retroviral vectors carrying four defined pluripotency genes (Oct4, Sox2, klf4, and c-Myc); however, recent advances have indicated that reprogramming can be accomplished using plasmids without a viral transfection system, or by using proteins passed into the cells through poly-arginine anchors without any genetic alteration of the adult cell (Okita et al., 2008; Zhou et al., 2009).
In the generation of iPS cells, the roles of the genes used for the reprogramming are crucial, as the success of the technique depends on the levels and patterns of expression of the factors used in transfection (Sridharan et al., 2009). Oct4 is a POU domain transcription factor that regulates genes downstream by binding to an octamer repeat sequence (Okamoto et al., 1990) and acts in conjunction with Sox2, a member of the Sox family of HMG box transcription factors (Yuan et al., 1995). Both factors play an essential role in the maintenance of self-renewal and pluripotency. An increase in Oct4 expression promotes mesoderm and endoderm formation, whereas the downregulation of either factor results in trophectoderm differentiation (Avilion et al., 2003; Niwa et al., 2000). Klf4 belongs to the Kruppel-like family of transcription factors and is a zinc-finger protein that can function both as a tumor suppressor and an oncogene (Foster et al., 20000; Katz et al., 2002). Another important protein, c-Myc, was one of the first proto-oncogenes found in human cancers (Dalla-Favera et al., 1982). The effects of c-Myc on chromatin structure enable Oct4 to activate or suppress target genes, whereas Klf4 may also function as a cofactor of Oct4 and Sox2 (Nakatake et al., 2006). Therefore, it seems likely that the relationships between these transcription factors could play critical roles in obtaining pluripotency and generating iPS cells (Knoepfler et al., 2006; Yamanaka, 2007). Although the importance of somatic cell reprogramming is considerable, it is still an experimental technology. Moreover, a recent report found that iPS cell-derived differentiating cells underwent early cellular senescence and had limited expansion potential (Feng et al., 2010).
The aim of this study was to examine whether established iPS cells exhibited molecular gene expression values as well as cellular characteristics comparable to control ES cells. To achieve this, we established several iPS cell lines from mouse embryonic fibroblasts (MEFs) using lentiviral vectors carrying the four defined pluripotency factors, Oct4, Sox2, c-Myc, and Klf4. We confirmed their typical ES-like cellular characteristics with respect to morphology, ES cell marker expression and in vitro or in vivo differentiation, using immunocytochemistry and histological analysis, and in addition, analyzed their global gene transcription profiles using DNA microarrays to compare them with control D3-ES cells and parental MEF cells. DNA microarray analysis is a powerful tool used in transcriptome profiling and the identification of stem cell characteristics (Ashton et al., 2007; Luo et al., 2006), and its use in the analysis of gene expression values for specific functional groups of genes will help to identify those iPS cells with true ES-like characteristics.
Materials and Methods
All chemicals and reagents were purchased from Sigma-Aldrich Corporation (St. Louis, MO, USA) unless otherwise stated.
MEF cell preparation
For MEF cell preparation, uteri isolated from 13.5-day pregnant C57BL/6 mice were washed with phosphate-buffered saline (PBS). The head and visceral tissues were removed from isolated embryos, and the bodies were washed with fresh PBS, minced using a pair of scissors, transferred into a solution of 0.25 mM trypsin/1 mM EDTA, and incubated at 37°C in a shaking incubator for 30 min. After trypsinization, the cells were dissociated by pipetting up and down, and were then collected by centrifugation. The pelleted cells were washed twice in culture medium and the resuspended cells were cultured on 100-mm dishes (≥1 × 106) in a 5% CO2 incubator at 37°C in Dulbecco's Modified Eagle's Medium (DMEM, Gibco, Grand Island, NY, USA) containing 10% defined-fetal bovine serum (FBS, Hyclone, Logan, UT, USA), 50 units/mL of penicillin, and 50 μg/mL of streptomycin. The third to fifth passages of cultured MEF cells were used for infection.
Lentiviral infection and cell culture
For the generation of iPS cells, 1 × 105 fibroblasts in each case were infected for 24 h in a suspended or adherent cell state with concentrated self-inactivating human immunodeficiency virus (HIV) type-1-based lentiviral vectors carrying the four transcription factors, Oct4/Sox2/Klf4, and c-Myc. The transcription factors were obtained by direct PCR of D3-ES cell cDNA. Adherent cell infection was carried out in 6-mL culture medium containing a concentrated solution of the four viral vectors. For the suspended cell infection, following trypsinization, detached MEF cells were collected by centrifugation, pelleted cells were resuspended in 0.5-mL culture medium together with approximately 0.5 mL of a concentrated solution containing the four viral vectors, and slowly mixed for 10 min, and then the MEF cell/virus complexes were plated on culture plates. The infection level was confirmed by the expression of the VENUS protein reporter gene, a variant of the yellow fluorescent protein gene. D3-ES cells and iPS cells were maintained on 10 μg/mL mitomycin C-treated STO cells (ATCC, Rockville, MD, USA) in ES cell culture medium composed of 80% DMEM medium, 20% defined-FBS, 1000 units/mL leukemia inhibitory factor (ESGRO: Chemicon International, Middlesex, UK), 1% nonessential amino acids (NEAA), and 0.55 mM β-mercaptoethanol (Gibco).
Stem cell marker staining
To examine stem cell marker expression, fifth to sixth passage cultured iPS cell lines were immunostained as follows: to detect alkaline phosphatase (AP) activity, iPS cell colonies were fixed in 4% paraformaldehyde (PFA) for 1 to 2 min and then treated with Fast Red Violet/Naphthol AS-BI phosphate mixed solution for 15 min (Millipore, Billerica, MA, USA). To detect mouse stem cell surface markers, fix iPS colonies for stage-specific embryonic antigens (SSEA-1), and for Oct-4 with PFA and permeabilized only the Oct-4 colonies with Triton X-100. To block any nonspecific binding, the cells were incubated first in 10% normal goat serum for 1 h, and then colonies were incubated with the primary antibody at 4°C overnight. After washing with PBS, cells were incubated with the secondary antibody for 1 h and nuclei were stained with 5 μg/mL DAPI. The primary antibodies used included a monoclonal antibody against SSEA-1 (1:50; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and a polyclonal antibody against Oct4 (1:250; Santa Cruz). The secondary antibodies used were goat antimouse tetramethylrhodamine isothiocyanate (TRITC, 1:200, Jackson Laboratories, West Grove, PA, USA) and goat antirabbit TRITC (1:200, Jackson Laboratories).
RT-PCR
For the RT-PCR analysis of stem cell marker expression or differentiation ability, we examined the 6th to 10th, or 14th to 18th passages of the cultured iPS cell lines, respectively. Total RNA was isolated using TRI reagent, according to the manufacturer's protocol (Sigma-Aldrich Company, Dorset, UK). Complementary DNA was synthesized from approximately 1 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen, Grand Island, NY, USA), and PCR was performed with AccuPrime DNA Taq polymerase (Invitrogen). The synthesized cDNA was amplified using 30 cycles of PCR with an annealing temperature of 52 to 60°C. The PCR products were size fractionated by 1% agarose gel electrophoresis and visualized by ethidium bromide staining. The final analysis was obtained using an image analyzer (Bio-Rad, Hercules, CA, USA).
Primer sequences for marker genes were: mOct4 endo (1061 bp: forward, 5′-gaattcccatggctggacacctg-3′; reverse, 5′-gcggccctcagtttgaatgcat-3′); mOct4 exo (1217 bp: forward, 5′-gaattcccatggctggacacctg-3′; reverse, 5′-atgctcgtcaagaagacagg-3′); mSox2 endo (962 bp: forward, 5′-gaattcgcatgtataacatgatg-3′; reverse, 5′-gcggccctcacatgtgcgacagg-3′); mSox2 exo (1118 bp: forward, 5′-gaattcgcatgtataacatgatg-3′; reverse, 5′-atgctcgtcaagaagacagg-3′); mC-Myc endo (1367 bp: forward, 5′-gaattcggctggatttcctttgg-3′; reverse, 5′-gcggcccttatgcaccagagtt-3′); mC-Myc exo (1524 bp: forward, 5′-gaattcggctggatttcctttgg-3′; reverse, 5′-atgctcgtcaagaagacagg-3′); mKlf4 endo (1427 bp: forward, 5′-gaattcacatggctgtcagcgac-3′; reverse, 5′-gcggcccttaaaagtgcctcttc-3′); mKlf4 exo (1583 bp: forward, 5′-gaattcacatggctgtcagcgac-3′; reverse, 5′-atgctcgtcaagaagacagg-3′); mCripto (423 bp: forward, 5′-ggtacttctcatccagtgtg-3′; reverse, 5′-tgaggtcctggtccatcacg-3′); mFGF4 (377 bp: forward, 5′-aaggcttcggcggctctact-3′; reverse, 5′-acagtctaggaaggaagtgg-3′); mNanog (449 bp; forward, 5′-tgagatgctctgcacagagg-3′; reverse, 5′-cagatgcgttcaccagatag-3′); and mzfp296 (410 bp: forward, 5′-cgacaccgacattgagatgc-3′; reverse, 5′-gcaacttccaaggactagtg-3′). Additional primer sequences for differentiation marker genes were: α-amylase (498 bp: forward, 5′-ggtctggaaatgaagatgaa-3′; reverse, 5′-cttggaaaatgaaaggtctg-3′); α-fetoprotein (525bp: forward, 5′-ggcaacaaccattattaagc-3′; reverse, 5′-gcaattcttcttccagattg-3′); β-enolase (494 bp: forward, 5′-ctgtggaacacatcaacaag-3′; reverse, 5′-ctcattgttctccaggatgt-3′); Rennin (495 bp: forward, 5′-gggctacacagctcttagaa-3′; reverse, 5′-gtagtggatggtgaagtcgt-3′); βIII tubulin (493 bp: forward, 5′-aaggtgcgtgaggagtatc-3′; reverse, 5′-gtagctgctgttcttgctct-3′); Map2 (485 bp: forward, 5′-tgaaactgaaccacagacaa-3′; reverse, 5’-agtgcttctcgtcatgtctt-3′), and G3PDH (450 bp: forward, 5′-gtcgtggagtctactggtgt-3′; reverse, 5′-gtcatcatacttggcaggtt-3′).
Real-time PCR quantification
For the semiquantitative real-time PCR analysis, we examined the 6th to 10th passages of the cultured iPS cell lines. Total RNA was extracted as described above and standard cDNA was synthesized using an oligo (dT) 12–18 primer and Superscript II reverse transcriptase (Invitrogen). Real-time RT-PCR was performed with endogenous (endo) or exogenous (exo) gene primer sets using a Chromo4 real-time thermal cycler (Bio-Rad,) and a DNA Engine Opticon 3 fluorescence detection system (MJ Research, Waltham, MA, USA) according to the manufacturer's instructions. In all experiments, G3PDH mRNA served as an internal standard. The threshold cycle (Ct) value represents the cycle number at which the sample fluorescence rose statistically above the background. To monitor the reactions, we followed the protocol described by the DyNAmo SYBR green qPCR kit, which contained a modified Tbr DNA polymerase, SYBR Green, optimized PCR buffer, 5 mM MgCl2, and a dNTP mix that included dUTP (Finnzyme Oy, Espoo, Finland). Gene expression was quantified using the 2-ddCt method (Livak and Schmittgen, 2001). Primer sequences for marker genes were mOct4 endo (215 bp: forward, 5′-ctagagaaggatgtggttcg-3′; reverse, 5′-tcaggaaaagggactgagta-3′), mOct4 exo (228 bp: forward, 5′-actcagtcccttttcctgag-3′; reverse, 5′-atgctcgtcaagaagacagg-3′), mSox2 endo (154 bp: forward, 5′-ggagtggaaacttttgtcc-3′; reverse, 5′-gggaagcgtgtacttatcct-3′), mSox2 exo (187 bp: forward, 5′-acggccattaacggcacact-3′; reverse, 5′-atgctcgtcaagaagacagg-3′), mC-myc endo (186 bp: forward, 5′-aggaagaaattgatgtggtg-3′; reverse, 5′-ctggatagtccttccttgtg-3′), mC-myc exo (234 bp: forward, 5′-acgtcacctctgaaaaggac-3′; reverse, 5′-atgctcgtcaagaagacagg-3′), mKlf4 endo (168 bp: forward, 5′-aaccttaccactgtgactgg-3′; reverse, 5′-aaaagtgcctcttcatgtgt-3′), mKlf4 exo (224 bp: forward, 5′-ctttcagtgccagaagtgtg-3′; reverse, 5′-atgctcgtcaagaagacagg-3′), and G3PDH (155 bp: forward, 5′-atggccttccgtgttcctac-3′; reverse, 5′-tggtcctcagtgtagcccaa-3′).
Western blot analyses
To examine stem cell protein expression, we analyzed the 19th and 20th passages of our cultured iPS cell lines. Antibodies against Oct 4 (H-134, Santa Cruz), Nanog (H-155, Santa Cruz), Sox2 (H-65, Santa Cruz), c-Myc (C-19, Santa Cruz), Klf4 (H-180, Santa Cruz), and β-actin (H-196, Santa Cruz) were used for Western blotting. To isolate proteins, iPS cells were lysed in lysis buffer (1% Triton X-100, 100 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10% glycerol, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 1 mM p-nitrophenyl phosphate, 1 mM PMSF). After the cells had been incubated on ice for 30 min, the lysates were centrifuged and the amount of protein in the cleared lysates was quantified. The protein samples (20 μg) were separated on 10 to 12% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane (0.2 mm Schleicher and Schuell Inc., Keene, NH, USA). After blocking with the TBS buffer (10 mM/l Tris-HCl pH 7.5, 150 mM/l NaCl) containing 5% nonfat dry milk and 0.1% Tween-20, the membrane was incubated with the primary antibodies, followed by incubation with goat antirabbit IgG peroxidase conjugates (Santa Cruz). The antibodies were detected using an enhanced chemiluminescence reagent (Amersham Bioscience, Piscataway, NJ, USA).
In vitro differentiation and immunofluorescence
To induce differentiation into the three embryonic layers, iPS cell colonies were cultured in suspension to form embryoid bodies (EBs) through an 8-day induction procedure that consisted of 4 days of culture as aggregates without 0.1 μM retinoic acid (RA), followed by 4 days of culture with RA (4 − /4+). EBs were cultured in differentiation medium (DMEM/F12 containing 1 mM glutamine, 0.55 mM β-mercaptoethanol, 1% NEAA, and 10% FBS). Cystic EBs were plated onto 0.1% gelatin-coated plates and cultured for 2 weeks. In vitro differentiation was assessed by indirect immunocytochemistry. Differentiated cells were fixed in 4% PFA for 15 min. Primary antibodies used were: mouse anti-β-tubulin III (Tuj1, 1:1000, Chemicon) as an ectoderm marker, mouse antisarcomeric-α-actinin (1:200) as a mesoderm marker, and mouse anti-α-fetoprotein (1:200, Santa Cruz) as an endoderm marker.
Teratoma formation and histological analysis
To analyze the formation of teratomas, 1 × 106 cells from the 14th to 18th passages of iPS cultures were suspended in 100 μL ES cell culture medium and inoculated beneath the testis capsule of a 5-week-old severe combined immunodeficient (SCID) mouse (BALB/c-nu Slc strain). Seven to 8 weeks after the injection, the resulting tumors were surgically dissected from the mice, fixed in PBS containing 4% PFA, and embedded in paraffin. Sections were stained with hematoxylin and eosin.
DNA microarray analysis
For microarray experiments, we used the 11th to 13th passages of our cultured iPS cell lines. Total RNA from D3-ES cells, six iPS cell lines, or MEF cells were extracted as described above and labeled with Cy3. For the preparation of cRNA targets for microarray analysis, 2 μg of total RNA were added to the reaction mix in a final volume of 12 μL, containing a T7 oligo dT primer, we then added 2 μL of 10 × first-strand buffer, 4 μL of 5 mM dNTP mix, 1 μL of RNase inhibitor (20 U/μL), and 1 μL of SuperscriptTM II RNase H-reverse transcriptase (200 U/μL) to make a final volume of 20 μL. To synthesize second-strand cDNA, 10 μL of 10 × second-strand buffer, 63 μL of nuclease-free water, 4 μL of 5 mM dNTP mix, 2 μL of DNA polymerase mix (20 U/μL), and 1 μL of RNase H (2 U/μL) were added to the 20 μL reaction mix. The double-stranded (ds) DNA was purified and the volume of the dsDNA solution was adjusted by drying the solution or by adding nuclease-free water to a final volume of 14 μL. For in vitro transcription, 4 μL of 10 × reaction buffer, 12 μL of ATP, CTP, and GTP mix (25 mM), 3 μL of UTP (50 mM), 3 μL of amino allyl UTP, and 4 μL of T7 enzyme mix was added to 14 μL of the dsDNA solution. The reaction mix was incubated for 14 h at 37°C followed by purification of the cRNA. The cRNA yield was quantified by measuring the UV absorbance at 260 nm, and 5 μg of cRNA for each sample was coupled with Cy-dye (Amersham Pharmacia, Uppsala, Sweden). Then the Cy-dye labeled cRNA (target) was fragmented for hybridization. In order to hybridize the fragmented target to an Agilent oligo microarray (G4121B), we added the fragmented target to the 2 × hybridization buffer from the Agilent In situ Hybridization kit. Hybridization solution was applied to the Agilent oligo microarray, and the microarray was hybridized for 16 h at 60°C. The microarray was washed with wash solution I (6 × SSC, 0.00005% Triton-X 102) for 10 min at room temperature and then with wash solution II (0.1 × SSC, 0.00005% Triton-X 102) for 10 min at 4°C. The microarray was scanned with a GenePix 4000B scanner (Axon Instruments, San Diego, CA, USA), and the scanned images were analyzed using GeneSpring GX software to optimize the signal intensity of the spots.
Results
Generation and characterization of iPS cells from MEF cells
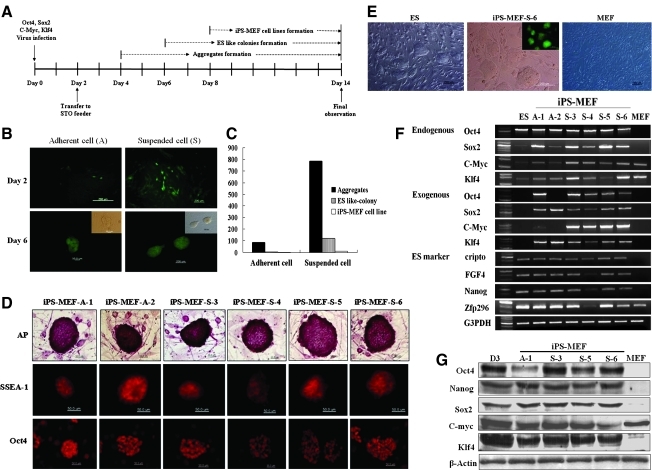
The four defined reprogramming factors Oct4, Sox2, c-Myc, and Klf4 were used to generate iPS cells from MEF cells, using the lentiviral infection method without antibiotic selection. As shown in the time line in Figure 1A, MEF cells were transferred onto STO feeder cells 2 days after infection and cultured in ES cell medium. We classified the morphological transformation of infected MEF cells into three types, aggregate formation, ES-like colony formation, and iPS-MEF cell line formation, up to 14 days after infection (Fig. 1A). In this study, MEF cells were infected using two different culture conditions, an adherent culture and a suspended culture (Fig. 1B). The infection level was detected easily by expression of the VENUS protein reporter gene (Figs. 1B, 1E, 3A, and Supplementary Fig. 1). As shown in Figure 1B, differences in infection levels between the two treatment groups were immediately apparent, with the suspended cell group showing higher levels of infection than the adherent cell group. When the infection efficiencies between the two treatment groups (adherent vs. suspended) were indicated numerically (Fig. 1C), the differences in aggregate formation, ES-like colony formation, and iPS-MEF cell line formation were 9.24 (785 vs. 85), 17.1 (120 vs. 7) and 5.0 (10 vs. 2)-fold higher, respectively, in the suspended cell group than in the adherent cell group. As shown in Supplementary Figure 1, typical ES cell-shaped iPS colonies appeared from day eight. From this experiment, we established 12 iPS cell lines, 2 of which came from the adherent cell group and 10 from the suspended cell group (Supplementary Fig. 2). The RT-PCR data indicated that the four exogenous or endogenous genes were expressed in all established iPS cell lines, although there were variations in expression levels.
FIG. 1.
Generation of induced pluripotent stem (iPS) cells from mouse embryonic fibroblast (MEF) cells using lentiviral vectors carrying four transcription factors. (A) Reprogramming protocol for MEF cells without drug selection. (B) Infected cells and embryonic stem (ES)-like colonies forming under different conditions, at 2 days and 6 days post infection, respectively. Fluoromicroscopic images of the cells indicate the expression of the Venus protein reporter gene. (C) Differences in infection efficiency according to cell treatment methods. (D) Verification of ES cell characteristics in generated iPS cell lines by examination of alkaline phosphatase activity, surface marker expression of SSEA-1, and Oct-4 expression. (E) Phase-contrast images of D3-ES cells, iPS colonies, and MEF cells. (F) RT-PCR analysis of expression of infected genes and ES marker genes in iPS cell lines. (G) Western blot analysis of gene expression in iPS cell lines.
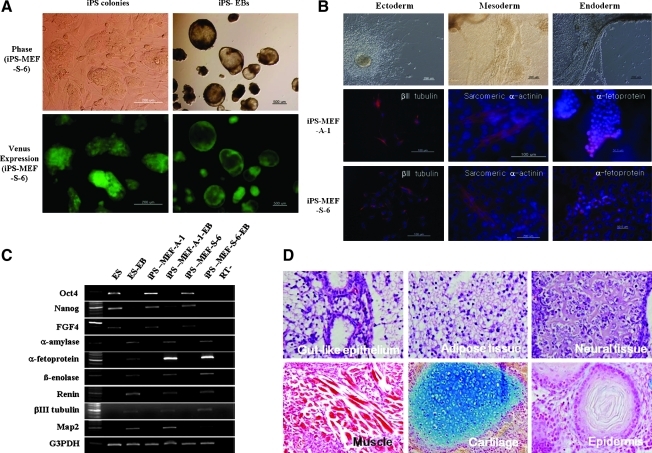
FIG. 3.
In vitro and in vivo differentiation of mouse iPS cells into the three germ layers. (A) Phase-contrast and fluorescent images of day four iPS-MEF-S-6 colonies cultured on STO feeder cells and their day eight embryoid bodies (EBs) prepared on a bacteriological plate. Fluoromicroscopic images of the cells indicate expression of the Venus protein reporter gene. (B) Immunofluorescence analysis of two differentiated iPS clones (iPS-MEF-A-1 and iPS-MEF-S-6) using cell markers for the three germ layers. (C) RT-PCR analysis of ES cell marker and differentiation marker expression in D3-ES cells, two different iPS cell lines, and their EBs. ES cell markers: Oct4, Nanog, and FGF4; endoderm markers: α-amylase and α-fetoprotein; mesoderm markers: β-enolase and Renin; ectoderm markers: βIII tubulin and Map2. (D) Histological examination of differentiated tissue structures found in teratomas formed in the testis of SCID mice following inoculation with mouse iPS cells. Endoderm tissue: gut-like epithelium; mesoderm tissue: adipose tissue, muscle and cartilage; ectoderm tissue; neural tissue and epidermis.
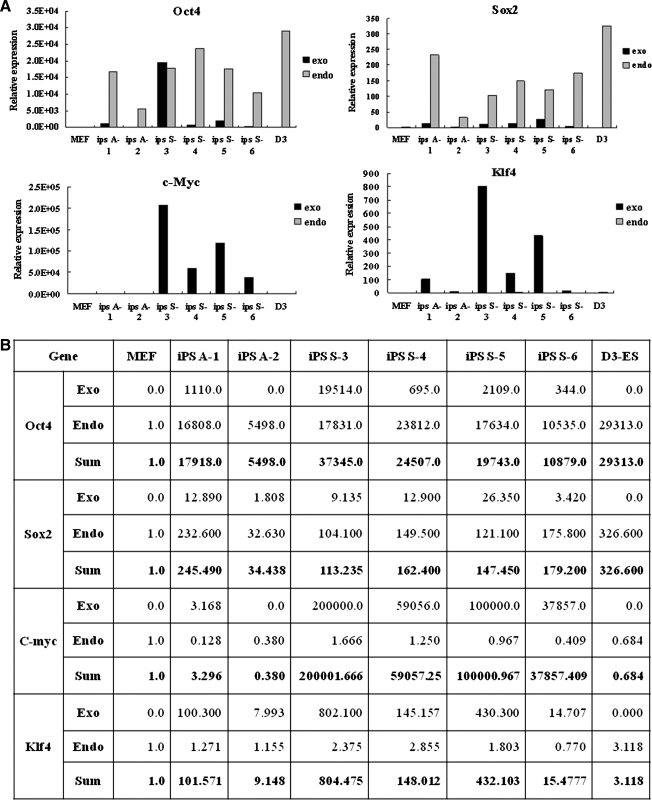
When we characterized six of the iPS cell lines (two from the adherent cell group: iPS-MEF-A-1 and iPS-MEF-A-2; and four from the suspended cell group: iPS-MEF-S-3, iPS-MEF-S-4, iPS-MEF-S-5, and iPS-MEF-S-6), as shown in Figure 1D, we found that alkaline phosphatase, SSEA-1, and Oct4 were highly expressed in all six cell lines. In addition, the iPS cells had transformed from their parental MEF cell morphology to the typical ES cell shape (Fig. 1E). The RT-PCR (Fig. 1F) analysis revealed that the four exogenous genes (Oct4, Sox2, c-Myc, and Klf4) were most strongly expressed in the reprogrammed iPS cells, although one of the cell lines from the adherent cell group, iPS-MEF-A-2, did not express Oct4 and c-Myc genes. Additionally, the endogenous copies of Oct4, Sox2, c-Myc, and Klf4 were all clearly expressed in iPS cells in contrast to MEF cells. On the other hand, Oct4 was clearly expressed in ES cells while Sox2, c-Myc, and Klf4 genes were expressed more weakly, and MEF cells expressed C-myc and Klf4 genes. Moreover, the pluripotency ES markers (crypto, FGF4, Nanog, and Zfp296) were all expressed in most iPS cells and were comparable to ES cells. Finally, Western blot analysis (Fig. 1G) revealed that iPS cells expressed the major stem cell marker proteins at similar levels to ES cells, while MEF cells exhibited C-myc protein expression. To examine the transduction levels of the four defined factors in iPS cells, we performed semiquantitative real-time PCR. As shown in Figure 2A, most genes showed high expression levels in the iPS cell lines derived from the suspended cell group (iPS-MEF-S). In particular, the exogenous c-Myc expression values were significantly higher than in the adherent iPS cell group (iPS-MEF-A). When the results were converted into numeric values (Fig. 2B), one adherent iPS cell line (iPS-MEF-A-1) presented a similar expression pattern to the control ES cells: Oct4 and Sox2 expression was high, whereas c-Myc and Klf4 expression was low. As mentioned above, we found that the exogenous Oct4 and c-Myc genes were silenced in iPS-MEF-A-2 cells.
FIG. 2.
Semiquantitative real-time PCR analysis of the expression levels of the four pluripotent-related genes in D3-ES cells, six iPS cell lines, and MEF cells. The results are expressed graphically (A) and numerically (B). MEF cells were used as a control to calculate the values of the other cell groups.
In vitro and in vivo differentiation of iPS cells
We examined the in vitro and in vivo differentiation of iPS cells to determine their pluripotent characteristics. As shown in Figure 3A, embryoid bodies formed well in suspension culture at 6 to 8 days. The EBs were then plated out and we confirmed that these cultured cells had differentiated into the three germ cell layers using immunocytochemistry (Fig. 3B) and RT-PCR (Fig. 3C). Markers for cell types from the three germ layers: neuronal cells (β-tubulin, ectoderm), cardiac cells (sarcomeric α-actinin, mesoderm), and hepatocytes (α-fetoprotein, endoderm) were all detected by immunocytochemistry in two representative iPS cell lines (iPS-MEF-A-1 and iPS-MEF-S-6). Furthermore, we used RT-PCR to show that these cells expressed genes from the three germ layers: endoderm, α-amylase, and α-fetoprotein; mesoderm, β-enolase, and rennin; and ectoderm, βIII tubulin, and Map2. However, the remaining four iPS cell lines (iPS-MEF-A-2, iPS-MEF-S-3, iPS-MEF-S-4 and iPS-MEF-S-5) showed weak EB formation and low differentiation ability. In order to examine the developmental pluripotency of the iPS cells, iPS-MEF-S-6 cells were inoculated beneath the testis capsule of SCID mice. Histological examination revealed that the teratomas contained representative tissues from the three germ layers. Differentiated tissues included gut-like epithelium (endoderm), adipose tissue, muscle, cartilage (mesoderm), neural tissue, and epidermis (ectoderm) (Fig. 3D).
Global gene expression analysis of iPS cells
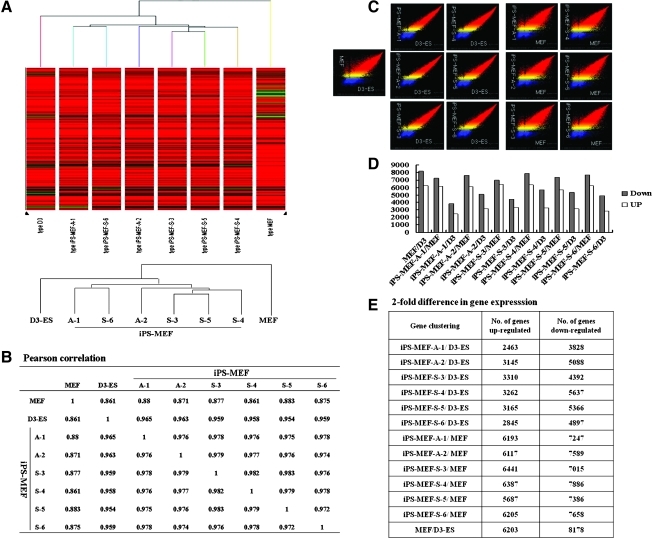
The global gene expression profiles of D3-ES cells, the iPS cell lines, and MEF cells were compared using DNA microarrays (Fig. 4), and we found that the transcriptome of the iPS cells was similar to that of the D3-ES cells. Hierarchical clustering and Pearson correlation analysis revealed that iPS cells (0.954 to 0.965) were clustered closely with D3-ES cells, but were distant from the parental somatic MEF cells (0.861 to 0.883) (Fig. 4A and B). Among the iPS cell lines, gene expression patterns for the iPS-MEF-A-1 and iPS-MEF-S-6 cell lines were most closely related to D3-ES cells. The scatter-plot presentations (Fig. 4C) also showed that gene expression patterns for the iPS cells were very tightly correlated to D3-ES cells and were as different from MEF cells as are D3-ES cells. When the twofold differences in gene expression between the cell groups were examined (Fig. 4D and E), the numbers of up- and downregulated genes in iPS cells compared to MEF cells were much greater (upregulated: 5687–6441; downregulated: 7015–8178) than for iPS cells against D3-ES cells (upregulated: 2463–3310; downregulated: 3828–5637). These results demonstrated that the gene expression patterns in iPS cells and D3-ES cells are more closely related to each other than they are to MEF cells.
FIG. 4.
Global gene expression analysis of iPS cells. (A) Hierarchical clustering of D3-ES cell line, six iPS cell lines (iPS-MEF-A-1, iPS-MEF-A-2, iPS-MEF-S-3, iPS-MEF-S-4, iPS-MEF-S-5, and iPS-MEF-S-6) and MEF cells. The transcriptome of the iPS clones is very similar to that of D3-ES cells. (B) Pearson correlations were calculated from the gene clustering results and showed close correlations between iPS cell groups and D3-ES cells. (C) Scatter-plot presentation of the expression values for all probe sets derived from genome-wide microarray data indicated that there were close correlations between iPS cell groups and D3-ES cells. (D–E) Examination of twofold differences in gene expression between cell groups indicated that fewer genes were up- or down-regulated in iPS cells compared to D3-ES cells than in iPS cells or D3-ES cells compared to MEF cells.
In addition, we analyzed the expression patterns of several pluripotency-related functional gene groups, such as stem cell proteins (413 genes), cell surface markers (141 genes), cell death proteins (598 genes), JAK–STAT signaling pathway proteins (161 genes), P13K–AKT signaling pathway proteins (219 genes), endothelial cell proteins (173 genes), cardiovascular cell proteins (173 genes), and neurogenesis and neural stem cell proteins (430 genes). A Treeview arrangement revealed similar gene expression patterns between iPS cell lines and D3-ES cells (Supplementary Fig. 3). In four of the functional groups, stem cell proteins, cell death proteins, cardiovascular cell proteins, and neurogenesis and neural stem cell proteins, there was a strong correlation between iPS-MEF-S-5 and D3-ES cells. We also found a strong correlation between iPS-MEF-S-6 and D3-ES cells in the ES signaling pathway proteins (JAK–STAT and P13K–AKT), and in the cell surface marker group, we observed a close relationship between iPS-MEF-A-1 and D3-ES cells (Supplementary Fig. 3).
With respect to the functional classification of the cell lines generated in this study, representatives of the highly expressed genes are listed numerically in Table 1. In the stem cell protein group, expression values for the following iPS cell line genes were lower than those in D3-ES cells: SRY-box–containing gene 2 (Sox2: ×2.58- to 10.0-fold; 99.532 to 387.660 vs. 1000.000); the POU domain class 5 transcription factor 1 (Pou5f1: ×1.6- to 2.2-fold; 226.941 to 313.312 vs. 500.000); the developmental pluripotency associated 5A gene (Dppa5a: ×1.04- to 1.41-fold; 236.813 to 321.208 vs. 333.333), and ES cell-expressed Ras (Eras, ×1.08- to 2.54-fold; 131.018 to 157.314 vs. 333.333). However, Lin28 (×1.43 to 2.33-fold; 285.450 to 466.461 vs. 200.000) and DNA methyltransferase 3b (Dnmt3b: ×1.33- to 2.64-fold; 60.613 to 120.153 vs. 45.455) showed higher expression values in iPS cells.
Table 1.
Functional Classification of Genes Highly Expressed in Mouse-Induced Pluripotent Stem (iPS) Cells
| |
|
Expression ratio (D3-ES or iPS-MEF/MEF) |
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein | Function | D3 | iPS-A1 | iPS-A2 | iPS-S3 | iPS-S4 | iPS-S5 | iPS-S6 | Gene bank accession no. |
| Stem cell proteins | |||||||||
| SRY-box containing gene 2 | Embryo development | 1,000.000 | 387.660 | 364.502 | 141.929 | 239.239 | 99.532 | 376.608 | NM_011443 |
| POU domain, class 5, transcription factor 1 | Undifferentiation | 500.000 | 313.312 | 271.949 | 226.941 | 249.762 | 244.215 | 231.695 | NM_013633 |
| Developmental pluripotency associated 5A | Pluripotency | 333.333 | 321.208 | 247.692 | 236.813 | 288.791 | 263.846 | 242.087 | NM_025274 |
| ES cell-expressed Ras | Proliferation | 333.333 | 310.046 | 147.962 | 157.314 | 139.212 | 131.018 | 133.703 | NM_181548 |
| Lin-28 homolog | Undifferentiation | 200.000 | 383.353 | 466.461 | 293.958 | 306.288 | 294.327 | 285.450 | NM_145833 |
| DNA methyltransferase 3B | De novo methylation | 45.455 | 91.264 | 94.406 | 120.153 | 60.613 | 83.793 | 65.900 | NM_010068 |
| Cell surface markers | |||||||||
| Platelet/endothelial cell adhesion molecule | Endothelial cell | 41.667 | 30.036 | 23.894 | 18.995 | 24.194 | 15.048 | 23.611 | NM_008816 |
| Dipeptidylpeptidase 4 | Immune regulation | 41.667 | 17.402 | 19.100 | 2.702 | 14.663 | 7.130 | 13.408 | NM_010074 |
| Tumor-associated calcium signal transducer 1 | Epithelial cell | 20.408 | 67.94 | 24.426 | 47.003 | 45.326 | 82.614 | 57.768 | NM_008532 |
| Cd68 antigen | Monocyte/macrophage | 10.753 | 1.440 | 2.914 | 2.617 | 4.414 | 2.250 | 4.231 | NM_009853 |
| Cd37 antigen | T-/B-cell interaction | 5.682 | 5.210 | 9.150 | 6.087 | 14.744 | 7.768 | 9.690 | NM_007645 |
| Cell death proteins | |||||||||
| Cell death-inducing DNA fragmentation factor αsubunit-like effector A | Activation of apoptosis | 142.857 | 49.755 | 43.066 | 29.827 | 59.000 | 40.066 | 60.555 | NM_007702 |
| Bcl2 modifying factor | Triggering apoptosis | 19.608 | 16.659 | 18.142 | 19.127 | 22.582 | 16.545 | 14.280 | BC079650 |
| Bcl2-associated athanogene 4 | Antiapoptotic | 8.197 | 4.939 | 3.563 | 3.338 | 4.403 | 3.079 | 5.307 | BC037239 |
| Bcl2-interacting killer | Inducing apoptosis | 5.917 | 7.786 | 8.333 | 8.927 | 8.900 | 6.006 | 13.567 | NM_007546 |
| Caspase recruitment domain family, member 10 | Apoptosis signaling | 4.255 | 6.994 | 11.859 | 9.359 | 9.895 | 8.183 | 5.718 | NM_130859 |
| JAK–STAT signaling pathway proteins | |||||||||
| Signal transducer and activator of transcription 4 | Activation | 37.037 | 34.246 | 22.491 | 21.688 | 35.185 | 23.266 | 31.777 | NM_011487 |
| Suppressor of cytokine signaling 2 | Suppression | 11.364 | 4.549 | 5.300 | 2.449 | 2.630 | 3.333 | 2.369 | AK033206 |
| Minichromosome maintenance deficient 5 | Cell cycle regulation | 7.194 | 5.150 | 4.590 | 4.675 | 4.404 | 4.172 | 4.370 | NM_008566 |
| Janus kinase 3 | Signal transduction | 2.653 | 1.661 | 1.756 | 1.333 | 1.129 | 1.024 | 2.001 | NM_010589 |
| Tyrosine kinase 2 | Cytokine signaling | 2.151 | 1.159 | 1.093 | 1.764 | 1.453 | 1.277 | 0.899 | NM_018793 |
| P13K–AKT signaling pathway proteins | |||||||||
| Inositol polyphosphate-5-phosphatase D | Signaling pathway | 125.000 | 37.851 | 31.037 | 38.925 | 40.333 | 25.533 | 32.962 | NM_010566 |
| Protein kinase C, zeta | Proliferation | 29.412 | 15.252 | 17.436 | 13.086 | 16.772 | 14.514 | 17.837 | NM_008860 |
| Ribosomal protein S6 kinase polypeptide 1 | Controlling cell growth | 5.556 | 7.463 | 8.535 | 9.852 | 8.086 | 8.467 | 6.659 | NM_009097 |
| Mitogen-activated protein kinase 8 | Proliferation | 4.545 | 2.880 | 1.414 | 1.856 | 2.644 | 1.660 | 3.954 | AK039129 |
| Cell division cycle 42 homolog | Regulation of cell cycle | 2.857 | 3.172 | 2.436 | 2.597 | 3.265 | 2.857 | 4.808 | L78075 |
| Others | |||||||||
| Integrin, alpha E, epithelial-associated | Adhesion | 29.412 | 16.198 | 17.134 | 12.643 | 20.812 | 12.877 | 24.561 | NM_008399 |
| Von Willebrand factor homolog | Blood homeostasis | 17.544 | 3.474 | 4.537 | 2.570 | 5.740 | 3.525 | 3.499 | AK078523 |
| Paired box gene 6 | Nervous system | 10.204 | 50.850 | 23.699 | 42.429 | 9.667 | 22.996 | 14.118 | AK045805 |
| Endothelial-specific receptor tyrosine kinase | Venous morphogenesis | 7.874 | 8.083 | 6.509 | 5.978 | 7.311 | 8.842 | 4.040 | NM_013690 |
| Oligodendrocyte transcription factor 2 | Neuron cell fate | 5.917 | 50.858 | 18.037 | 43.098 | 14.956 | 30.271 | 14.981 | NM_016967 |
| Fibrinogen, gamma polypeptide | Adhesion | 5.051 | 7.940 | 5.700 | 3.923 | 3.153 | 2.794 | 12.512 | NM_133862 |
| Nerve growth factor receptor | Neuron growth | 4.098 | 15.479 | 14.83 | 27.396 | 9.019 | 20.744 | 7.500 | NM_033217 |
| Calcium channel, voltage-dependent, N-type,alpha 1B subunit | Calcium entry | 2.793 | 1.964 | 2.204 | 2.764 | 3.004 | 1.195 | 3.191 | AK081230 |
| Troponin I, cardiac | Actin–myosin interaction | 2.119 | 1.406 | 2.194 | 2.642 | 2.192 | 1.987 | 2.518 | NM_009406 |
In the cell surface marker protein group, expression values of platelet/endothelial cell adhesion molecule 1 (Pecam1: ×1.39- to 2.77-fold; 15.048 to 30.036 vs. 41.667); dipeptidylpeptidase 4 (Dpp4: ×2.18- to 15.4-fold; 2.702 to 19.100 vs. 41.667) and Cd68 antigen (×2.44- to 7.47-fold; 1.440 to 4.414 vs. 10.753) were also lower in iPS cell lines than in D3-ES cells, while high expression ratios were detected for Tacstd 1 (×1.19- to 4.05-fold; 24.426 to 82.614 vs. 20.408) and Cd37 (×1.07- to 2.59-fold; 6.087 to 14.744 vs. 5.682) antigens in the iPS cell lines.
In the cell death protein group, the values of cell death-inducing DNA fragmentation factor α subunit-like effector A (Cidea: ×2.36- to 4.79-fold; 29.827 to 60.555 vs. 142.857) and Bcl2-associated athanogene 4 (Bag4: ×1.63- to 2.66-fold; 3.079 to 5.307 vs. 8.197) were lower in iPS cell lines than in D3-ES cells, whereas those of the Bcl2-interacting killer (Bik: ×1.02- to 2.29-fold; 6.006 to 13.567 vs. 5.917) and of the caspase-recruitment domain family, member 10 (Card10: ×1.34- to 2.79-fold; 5.718 to 11.859 vs. 4.255) were higher in iPS cell lines.
In contrast, in the ES cell signaling-related JAK-STAT pathway protein group, most of the gene expression ratios (Stat4, Socs2, Mcm5, Jak3, and Tyk2) were higher in D3-ES cells than in the iPS cell lines. In the cell proliferation-related p13K–AKT signaling pathways, the values of inositol polyphosphate-5-phosphatase D (Inpp5d: × 3.1- to 4.89-fold; 25.533 to 40.333 vs. 125.000); protein kinase C, zeta (prkcz: ×1.65- to 2.17-fold; 13.086 to 17.436 vs. 29.412); and mitogen activated protein kinase 8 (Mapk8; ×1.15- to 3.21-fold; 1.414 to 3.954 vs. 4.545) were lower in iPS cell lines than in D3-ES cells, whereas the expression of ribosomal protein S6 kinase polypeptide 1 (Rps6ka1: ×1.20- to 1.77-fold; 6.659 to 9.852 vs. 5.556) was higher in iPS cells.
In other differentiation protein groups, the expression ratios of endothelial-specific receptor tyrosine kinase (Tek), calcium channel voltage-dependent N type alpha 1B subunit (Cacna1b), and troponin I cardiac (Tnni3) in iPS cell lines were similar to those in D3-ES cells, whereas the expression ratios of the neurogenesis-related paired box gene 6 (Pax6: ×1.38- to 4.98-fold; 9.667 to 50.850 vs. 10.204), oligodendrocyte transcription factor 2 (Olig 2: × 2.53- to 8.59-fold; 14.956 to 50.858 vs. 5.917), and nerve growth factor receptor (Ngfr: ×1.83- to 6.69-fold; 7.500 to 27.396 vs. 4.098) were very highly expressed in iPS cell lines compared to D3-ES cells.
Discussion
This study revealed differences in the expression profiles of a number of stem cell-related functional genes between our iPS cells and D3-ES cells. On the other hand, we found that cellular characteristics such as morphology and expression markers in the iPS cells appeared similar to ES cells. With respect to their morphology, our established iPS cells appeared like typical ES cells, growing as compact, domed colonies with well-defined edges. Stem cell markers such as Oct4, SSEA1, and alkaline phosphatase were expressed homogeneously in iPS cells at levels comparable with the control D3-ES cells, as assessed by immunocytochemistry. In addition, the iPS cells expressed most ES markers, including Oct4, Sox2, c-Myc, Klf4, Nanog, crypto, FGF4, and Zfp296, as assessed by RT-PCR and Western blot analysis.
Using real-time PCR, we observed variations in the expression levels of the four pluripotency-related genes among the iPS cell lines examined and found that the suspended cell infection method (iPS-MEF-S) was more efficient than adherent culture (iPS-MEF-A) for the introduction of genes. Among the six iPS cell lines examined, two of the cell lines (iPS-MEF-A-1 and iPS-MEF-S-6) exhibited markers characteristic of differentiation into the three germ layers, both in vitro and in vivo, including neural, cardiac and hepatic cells in vitro, and gut-like epithelium, adipose tissue cells, neural tissue cells, muscle cells, cartilage cells, and epidermis in vivo. In addition, one of these cell lines produced a number of beating cardiac masses from cardiac cell differentiation and individual contracting cells exhibited cardiac-specific current recordings for potassium, sodium, and calcium channel analysis (data not shown). In contrast, the other four iPS cell lines showed weak EB formation and low differentiation ability, irrespective of marker expression and gene expression levels. From these results, it appeared that the iPS cell lines exhibited variations in stem cell stability, and thus further analysis is needed to assess their pluripotency. However, the global gene transcription analysis revealed significant differences in expression of the core pluripotency genes (Oct4/Sox2: × 2- to 10-fold lower) in our iPS cells compared to D3-ES cells, and it is possible that these differences may explain the weak pluripotency of iPS cells.
In this study, 12 iPS cell lines were established by reprogramming MEF cells using lentiviral vectors containing a quartet of transcription factors (Oct4, Sox2, c-Myc, and Klf4). We did not use drug selection and performed the infection of MEF cells in an adherent (A) or suspended (S) state. The infection efficiencies (A; 0.085%, S; 0.785%) were comparable to those reported by other groups (Okita et al., 2007; Wernig et al., 2007). A suspension environment provides three-dimensional spaces for the infected cells, which leads to an increase in the transduction efficiency during the two to 3 h in which the cells are suspended in medium before attachment. Thus, the suspended cell infection method resulted in a significantly higher reprogramming efficiency (9.24-fold) of MEF cells compared to the adherent cell infection method. We obtained 10 iPS cell lines by suspended infection, whereas the remaining two iPS cell lines were generated by adherent infection.
The roles played by the reprogramming genes used for the generation of iPS cells are crucial. Success may depend on the levels and patterns of expression of the factors used in infection. Analysis of real-time PCR revealed that Oct4 expression ratios were much higher in transformed iPS cell lines compared to parental MEF cells. However, Oct4 expression ratios varied among the iPS cell lines and the values were lower than in D3-ES cells. The level of Oct4 expression is an important determinant of cell fate in ES cells and it is known that other transcription factors might have direct effects on the increase in Oct4 expression (Nakatake et al., 2006; Yamanaka, 2007). Our results revealed that the endogenous Oct4 expression values showed a remarkable increase in iPS cells (×5,498- to 23,812-fold) compared to the parental MEF cells after infection with the four reprogramming factors, whereas most of the exogenous Oct4 expression values (0–19,514) were lower than the endogenous values. This means that the other transduction factors, Sox2, c-Myc, and Klf4, are practically affected by the increase in Oct4 expression levels during the generation of iPS cells. It is problematic that the endogenous c-Myc expression value was significantly higher in the cells derived from suspended cell infection than in those derived from adherent cell infection. Nevertheless, we cannot disregard the possibility that c-Myc may act on chromatin structure to enable the activation of Oct4.
According to the numeric values from the real-time PCR analysis, the exogenous c-Myc expression values of the two adherent iPS cell lines were significantly lower than the suspended iPS cell lines and their infection efficiency was also very low. It is interesting that iPS-MEF-A-1 showed a similar expression pattern to control ES cells: high Oct4 and Sox2 expression and low c-Myc and Klf4 expression. Furthermore, the exogenous Oct4 and c-Myc genes were silenced in the iPS-MEF-A-2 cell line. From our results, it is possible that adherent infection may provide the means for a more stable introduction of genes, although at a low infection efficiency. However, the use of integrating lentiviral vectors to generate iPS cells is always accompanied by problematic residual transgene expression, which may alter the expression of endogenous genes. Nevertheless, our karyotype analysis demonstrated that four suspended iPS cell lines [iPS-MEF-S-3(40, XX), iPS-MEF-S-4(40, XX), iPS-MEF-S-5(40, XX), iPS-MEF-S-6(40, XY)] have normal karyotype, whereas one adherent iPS cell line is normal [iPS-MEF-A-1 (40, XX)] and another adherent iPS cell line presents mosaicism [iPS-MEF-A-2 (40, XY; 40, XY, i(16)(A)] (Supplementary Fig. 4). It is very difficult, therefore, to determine the best conditions for infection.
From our results, there was no obvious relationship between infection efficiency and pluripotency of iPS cells. The overexpression of one particular transcription factor does not make a reprogrammed somatic cell pluripotent, and it is important that a balance is maintained among the four transcription factors to obtain pluripotency in generated iPS cells (Knoepfler et al., 2006; Yamanaka, 2007). Oct4 has a biphasic effect; an intermediate Oct4 level maintains pluripotency by stimulating Nanog and antagonizing Cdx2 (Chickarmane and Peterson, 2008) and a high level of Oct4 expression further suppresses Cdx2 and seems to stimulate Gata6, which in turn suppresses Nanog (Niwa et al., 2000). A >50% increase in the level of Oct4 can convert ES cells into extra-embryonic endoderm-like stem cells. Moreover, a transient increase in the level of Oct4 may play a role in mesoderm and neuroectoderm specification (Shimozaki et al., 2003; Zeineddine et al., 2006).
Although RT-PCR analysis revealed the expression of several genes associated with the pluripotency of ES cells, global messenger RNA expression analysis allowed us to perform large-scale analysis of gene expression in D3-ES cells, parental MEF cells, and their reprogrammed iPS cells at the same time. DNA microarray analysis is a potent tool for the measurement of transcriptome profiling for tens of thousands of genes and enables the identification of stem cell characteristics. On a large scale, clustering analysis revealed a high degree of similarity among the reprogrammed iPS cells (iPS-MEF-A-1, iPS-MEF-A-2, iPS-MEF-S-3, iPS-MEF-S-4, iPS-MEF-S-5, iPS-MEF-S-6), which were clustered closely with D3-ES cells, and were distant from the parental MEF cells, as determined by the Pearson correlation.
Among the 32,335 genes analyzed, a number of genes (upregulated: 5687–6441; downregulated: 7015–8178) showed a greater than twofold difference in expression in iPS cell lines compared to MEF cells, while 6203 (upregulated) and 8178 (downregulated) genes showed a greater than twofold difference in expression in D3-ES cells compared to MEF cells. In contrast, fewer genes (upregulated: 2463–3310; downregulated: 3828–5637) showed a greater than twofold difference in expression in iPS cell lines compared to D3-ES cells. These results demonstrate that our established iPS cells were more closely related to D3-ES cells at the global transcription level than to MEF cells, even when taking into account their different genetic origins: iPS cells (C57BL/6) and D3-ES cells (129S2/SvPas). However, it is worth noting that more genes were downregulated in iPS cells than were upregulated, compared to D3-ES cells. Among the large number of genes profiled, the more prominently upregulated genes in D3-ES cells, or iPS cell lines, were mostly stem cell factors such as Trap1a (Tumor rejection antigen P1A), Sox2, Oct4, Nanog, Dppa5a, E-ras, Gdf3 (Growth differentiation factor 3), and Zfp296 (Zinc-finger protein 296). Nevertheless, the expression values of these genes in iPS cells were significantly, or slightly, lower than those in D3-ES cells, whereas Lin28 and Alpl (alkaline phosphatase) expression values were higher in iPS cells than in D3-ES cells. According to our results, the levels of Oct4 and Sox2 in iPS cells were significantly lower (×1.6- to 2.2-fold and 2.58- to 10.0-fold, respectively) than in D3-ES cells, which may be the cause of their weak pluripotency. It is worth considering a report that showed that although pluripotency-related genes were more abundantly expressed in the 129 strain than in the C57BL/6 strain, the expression of the canonical pluripotency genes (Pou5f1, Nanog, and Sox2) required to maintain a normal ES cell-like phenotype showed no differences between the two strains (Sharova et al., 2007). We felt that it was very important to assess the ES-like iPS cells because their core pluripotency gene expression values differed from those of ES cells.
In this study, we analyzed the gene expression patterns of several pluripotency-related functional groups such as stem cell-specific proteins, cell surface markers, proteins involved in cell death, members of the JAK–STAT and P13K–AKT signaling pathways, endothelial cell-specific proteins, and proteins specific to cardiovascular cells and neurogenesis and neural stem cells. On a large scale, a Treeview arrangement revealed that there were very similar gene expression patterns in each functional group between iPS cells and D3-ES cells, in contrast to the parental MEF cells, although there were differences in individual gene expression values and the numbers of downregulated genes were high in iPS cells.
In the cell surface marker protein group, representative transcription factors such as Pecam1, Dpp4, Tacstd1, Vwf, Tek, Lag3, Itga3, Icam2, Nos3, Cd8a, Cd22, Cd37, Cd68, Cd79b, and Cd96 were highly expressed in all iPS cells, although their expression values were lower than in D3-ES cells. High expression ratios were also detected for Tacstd 1 (epithelial cell marker) and Cd37 antigen (T-/B-cell interaction marker) in iPS cell lines. In the cell death protein group, representative transcription factors such as Cidea, Cidec, Inpp5d, Bmf, Tcf7, Pycard, Spn, Camk1d, Bag4, C8b, C8g, Bik, Bmf, Dapk2, Dedd2, Dido1, Casp8ap2, Mbd4, Bcl2l14, Bfar, Pdcd7, and Dffb were highly expressed in both iPS cells and D3-ES cells. Of these, gene expression values for Cidea (apoptosis activation factor) and Bag4 (antiapoptotic factor) were lower in iPS cell lines than in D3-ES cells, whereas values for Bik (apoptosis inducing factor) and Card10 (apoptosis signaling factor) were higher in iPS cell lines. It was estimated that a greater number of apoptosis-related genes were expressed in iPS cells than in D3-ES cells. Also, this result may be related to the lower growth potential of iPS cells compared to D3-ES cells; the iPS cell lines examined showed a mean of 15 passages of viable subculture (10–23 passages). This result was confirmed in another study (Feng et al., 2010), which reported that iPS cells were capable of generating several types of differentiated cells with phenotypic and morphological characteristics similar to ES cells, but with a dramatically decreased efficiency. Their iPS cells also underwent early cellular senescence and showed limited expansion potential.
In addition, in the ES signaling-related JAK–STAT pathway, most of the gene expression ratios (Stat4, Socs2, Mcm5, Pias2, Jak3, and Tyk2) were higher in D3-ES cells than in iPS cell lines. This result may be related to the lower expression values of pluripotent genes in iPS cells than in D3-ES cells. In the cell proliferation-related p13K–AKT signaling pathway, representative transcription factors such as Inpp5d, Prkcz, Mapk8ipl, Rps6ka1, Mapk8, Raf1, Foxo1, and Prkcb1 were highly expressed in both iPS cells and D3-ES cells. In other differentiation protein groups, endothelial cell-specific markers such as Pecam1, Vwf, Tek, Ocln, Icam2, Nos3, Coll8a1, Cradd, Sod1, and Birc2; cardiovascular disease markers such as Itgae, Apoc1, Itga3, Itga6, Apoe, Ltb, Itgb4, Itgb7, Fgg, Mrpl15, Cacna1b, and Tnni3; and neurogenesis and neural stem cell markers such as Otx2, Pax6, Dmbx1, Zic2, Neurod1, Olig2, Sip1, Ngfr, Olig1, Bai2, Gfra3, Ntrk2, and Nrxn2 were highly expressed in both iPS cells and D3-ES cells. In particular, several neurogenesis markers, such as Pax6, Olig 2, and Ngfr genes, were very highly expressed in iPS cell lines compared to D3-ES cells. From these results, we expect that the numerical evaluation of gene expression using DNA microarrays will be an important means of assessing the characteristics of newly generated iPS cells.
Taken together, these results demonstrate that our established iPS cell lines present cellular morphological characteristics almost typical of ES cells with a clear correlation between global gene transcription in iPS cells and ES cells, as reported in other studies (Maherali et al., 2007; Takahashi and Yamanaka, 2006). However, there were variations in differentiation ability, regardless of marker expression and gene expression level. Although the most highly expressed genes were pluripotent genes, in iPS cells, the numbers of downregulated genes were greater than those of the upregulated genes compared to D3-ES cells, and many of the gene expression values in iPS cells were not comparable to those in D3-ES cells. Moreover, we found variation in the gene expression ratios among the different iPS cell lines. Nevertheless, the generation efficiency of iPS cells is very close to that of other stem cell types, and it is thought that the generation of iPS cells may soon become an alternative to the use of ES cells or somatic cell nuclear transfer ES cells in patient-specific cell therapies. The current goal in the generation of iPS cells is to develop a more stable and more efficient transfection method.
This study has demonstrated differences in the expression profiles of a number of stem cell-related functional genes between our iPS cells and ES cells, whereas their cellular characteristics, including morphology and marker expression, appeared similar to ES cells. These differences and similarities may affect their use in various applications; therefore, when characterizing iPS cells, in addition to cellular characterization, genome-based DNA microarray analysis may be required to identify whether iPS cells show true stem cell stability.
Supplementary Material
Acknowledgments
This study was supported by a grant (308008-5) from the Technology Development Program for Agriculture and Forestry, Ministry for Agriculture, Forestry and Fisheries, Republic of Korea, and the Agenda Program (No. PJ007577201003), Rural Development Administration, Republic of Korea. We thank Dr. Hiroyuki Miyoshi of the RIKEN BioResource Center for the lentiviral system.
Author Disclosure Statement
The authors declare that there are no conflicting financial interests.
References
- Ashton R.S. Peltier J. Fasano C.A., et al. High-throughput screening of gene function in stem cells using clonal microarrays. Stem Cells. 2007;25:2928–2935. doi: 10.1634/stemcells.2007-0468. [DOI] [PubMed] [Google Scholar]
- Avilion A.A. Nicolis S.K. Pevny L.H., et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane V. Peterson C. A computational model for understanding stem cell, trophectoderm and endoderm lineage determination. PLoS One. 2008;3:e3478. doi: 10.1371/journal.pone.0003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R. Bregni M. Erikson J., et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q. Lu S.J. Klimanskaya I., et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;12 doi: 10.1002/stem.321. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Foster K.W. Frost A.R. McKie-Bell P., et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- Katz J.P. Perreault N. Goldstein B.G., et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler P.S. Zhang X.Y. Cheng P.F., et al. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo Y. Schwartz C. Shin S., et al. A focused microarray to assess dopaminergic and glial cell differentiation from fetal tissue or embryonic stem cells. Stem Cells. 2006;24:865–875. doi: 10.1634/stemcells.2005-0392. [DOI] [PubMed] [Google Scholar]
- Maherali N. Sridharan R. Xie W., et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Nakatake Y. Fukui N. Iwamatsu Y., et al. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol. Cell Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. Miyazaki J. Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H. Masui S. Chambers I., et al. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell Biol. 2002;22:1526–1536. doi: 10.1128/mcb.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K. Okazawa H. Okuda A., et al. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- Okita K. Ichisaka T. Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K. Nakagawa M. Hyenjong H., et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Park I.H. Zhao R. West J.A., et al. Reprogramming of human somatic cells to pluripotency with defind factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Sharova L.V. Sharov A.A. Piao Y., et al. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev. Biol. 2007;307:446–459. doi: 10.1016/j.ydbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozaki K. Nakashima K. Niwa H, et al. Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development. 2003;130:2505–2512. doi: 10.1242/dev.00476. [DOI] [PubMed] [Google Scholar]
- Sridharan R. Tchieu J. Mason M.J., et al. Role of the murine reprogramming factors in the induction of pluripoptency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Tanabe K. Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defind factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson J.A. Itskovitz-Eldor J. Shapiro S.S., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wernig M. Meissner A. Foreman R., et al. In vitro reprogramming of fibroblasts into a pluripotent ES cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and now developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Yu J. Vodyanik M.A. Smuga-Otto K., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007 Nov;19 doi: 10.1126/science.1151526. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Yuan H. Corbi N. Basilico C., et al. Developmentalspecific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Zeineddine D. Papadimou E. Chebli K., et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev. Cell. 2006;11:535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Zhou H. Wu S. Joo J.Y., et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.