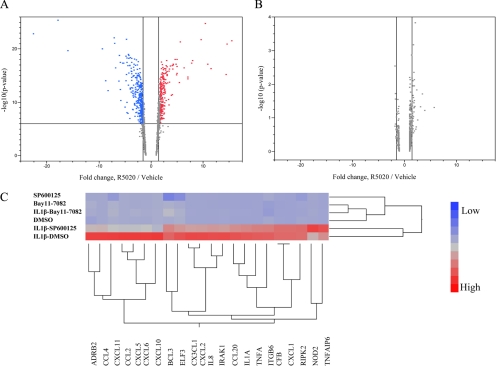
Figure 1.
Induction profiles of genes with putative NF-κB1/NF-κB-binding sites by R5020. A, Genes significantly regulated by R5020 in hMECs are analyzed for the presence of putative NF-κB1/NF-κB-binding elements in their regulatory sequences as described in Supplemental Data. Five hundred twenty-two genes were identified. Their regulation in hMECs transiently overexpressing hPR-B was assessed by two-way plotting of induction/repression levels vs. −log10 (P value) for corresponding probesets (A). B, Similar plot for T47D cells (primary expression data courtesy of Dean Edwards, Baylor College of Medicine). Vertical dashed lines represent 1.5-fold induction/repression boundaries. Horizontal dashed line in A represents Holm-adjusted statistical significance cutoff (6.02). None of the genes expressed in T47D cells passed the significance cutoff (α = 0.05) when adjusted for multiple comparisons. C, Dendogram showing the expression profile of 22 NF-κB and inflammatory genes in cells treated with vehicle (dimethylsulfoxide), a NF-κB inhibitor (Bay11-7082), or a JNK inhibitor (SP600125) with and without IL-β. The profiles were analyzed with the Ward hierarchical clustering algorithm using standardized data.