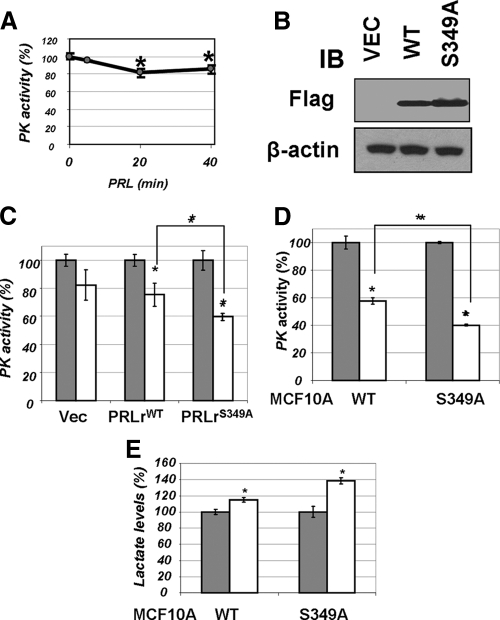
Figure 3.
A, Pyruvate kinase activity was determined in lysates (50 μg) from 293T cells treated with PRL (100 ng/ml for indicated time points) by a coupled enzymatic-based spectrophotometric assay as described in Materials and Methods. Average data from four independent experiments (each in triplicate) are presented as percent of activity measured in cells that did not receive PRL (±sd). Asterisks denote statistical significance of obtained differences (P < 0.05 by Student’s t test). B, Transient expression of indicated Flag-tagged PRLr species in 293T cells was verified by immunoblotting (IB) using indicated antibodies. C, Activity of pyruvate kinase was determined (as in panel A) in lysates from 293T cells transfected with vector control (pCDNA3) or vectors for expression of PRLr [wild type (WT) or S349A mutant] and treated (100 ng/ml PRL for 20 min, white bars) or not (gray bars) with PRL. Here and in similar subsequent figures, the average data from three independent experiments (each in triplicate) are presented as percent of activity of nontreated control cells (±sd). Absolute values of PKM activity in untreated cells transfected with different PRLr constructs were comparable (data not shown). Asterisks denote statistical significance of obtained differences (P < 0.05 by Student’s t test). D, Pyruvate kinase activity was measured in the lysates from MCF10a-derived cells expressing wild-type or S349A mutant of PRLr (described in details in Ref. 12) as in panel B. E, Lactate levels in the lysates from cells used in panel C were determined using a fluorescence-based lactate measurement assay. Asterisks signify that the difference in lactate levels between the treated and untreated samples is significant as determined by Student’s t test (P < 0.05).