Abstract
Diversity among Conus toxins mirrors the high species diversity in the Indo-Pacific region, and evolution of both is thought to stem from feeding-niche specialization derived from intra-generic competition. This study focuses on Conus californicus, a phylogenetic outlier endemic to the temperate northeast Pacific. Essentially free of congeneric competitors, it preys on a wider variety of organisms than any other cone snail. Using molecular cloning of cDNAs and mass spectrometry, we examined peptides isolated from venom ducts to elucidate the sequences and post-translational modifications of two eight-cysteine toxins (cal12a and cal12b of type 12 framework) that block voltage-gated Na+ channels. Based on homology of leader sequence and mode of action, these toxins are related to the O-superfamily, but differ significantly from other members of that group. Six of the eight cysteine residues constitute the canonical framework of O-members, but two additional cysteine residues in the N-terminal region define an O+2 classification within the O-superfamily. Fifteen putative variants of Cal12.1 toxins have been identified by mRNAs that differ primarily in two short hypervariable regions and have been grouped into three subtypes (Cal12.1.1–3). This unique modular variation has not been described for other Conus toxins and suggests recombination as a diversity-generating mechanism. We propose that these toxin isoforms show specificity for similar molecular targets (Na+ channels) in the many species preyed on by C. californicus and that individualistic utilization of specific toxin isoforms may involve control of gene expression.
Keywords: sodium channels, conotoxins, peptides, peptidomics
INTRODUCTION
Marine cone snails inject prey with paralytic peptide toxins, and more than 500 extant Conus species produce an enormous number of unique Cys-rich conotoxins (Gray et al., 1988; Norton and Olivera, 2006; Rockel et al., 1995). Arrangements of Cys residues and internal Cys–Cys disulfide bonds define a large number (more than 20) of particular structural frameworks, with most sequences falling into eight categories (Aguilar et al., 2005; Möller et al., 2005; Norton and Olivera, 2006; Pi et al., 2006). Conotoxin precursors contain a hydrophobic leader (signal) sequence and pro-peptide region, both of which are removed by proteolysis during biosynthesis, and the primary structure of the signal sequence is generally highly conserved between toxins having identical Cys-frameworks. Although the combination of leader sequence and Cys-framework has been used to define at least 15 superfamilies (Olivera and Cruz, 2001; Terlau and Olivera, 2004), the leader sequence is of primary importance (Halai and Craik, 2009). Many conotoxins also show a wide variety of post-translational modifications (PTMs), which further increases toxin diversity (Craig et al., 1999; Olivera, 1997).
Peptides within a superfamily generally target different members of a specific functional class of protein (McIntosh and Jones, 2001). For example, μ-type toxins (M-superfamily, Cys-framework 3) and μO-toxins (O-superfamily, framework 6) preferentially block different isoforms of voltage-gated Na+ channels in nerve and muscle (Terlau and Olivera, 2004; Daly et al., 2004; McIntosh et al., 1995). In general, the amino acid composition of intra-Cys loops of such closely related toxins is thought to confer specificity towards a high-affinity molecular target (McIntosh and Jones, 2001; Norton and Olivera, 2006; Woodward et al., 1990).
Diversity in toxin structure and specificity is reflected in the feeding behavior of different Conus species. Almost all species prey on one general type of organism, e.g. vermiforms (polychaetes or hemichordates), molluscs or fish, with many preying on specific taxa within these groups (Rockel et al., 1995). Interspecific competition within the genus is believed to have been an important evolutionary force leading to feeding-niche specialization and toxin diversification (Conticello et al., 2001; Duda et al., 2001; Duda and Palumbi, 1999; Espiritu et al., 2001). These ideas are primarily based on studies of Conus species from the Indo-Pacific region, an area with the greatest species diversity (Kohn, 1998). Comparatively few conotoxins have been identified from eastern Pacific (Hopkins et al., 1995) or Atlantic species (Luna-Ramirez et al., 2007; Möller et al., 2005), but both regions contain endemic species with novel toxins that fit into the conceptual framework described above.
Conus californicus Hinds 1844, arguably the most unusual member of the genus, is endemic to the temperate northeast Pacific coast. Over most of its range (central California to southern Baja California), it is the only representative of the genus, a situation that may have existed since the Miocene (Stanton, 1966). Phylogenetic studies indicate an extremely distant relationship between C. californicus and the rest of the genus, including eastern Pacific members (Duda and Kohn, 2005; Duda et al., 2001; Duda and Palumbi, 2004; Espiritu et al., 2001). Most importantly, C. californicus is a generalist feeder with the broadest diet of any Conus species. It preys primarily on molluscs and worms, with gastropods and bivalves being the major prey types in one reported study of gut contents (Kohn, 1966). Octopus and fish are also consumed (Kohn, 1966; Stewart and Gilly, 2005). Although such a diverse diet suggests that C. californicus may express novel toxins, this species has been largely overlooked (Cottrell and Twarog, 1972; Elliott and Kehoe, 1978; Elliott and Raftery, 1979; Whysner and Saunders, 1963; Whysner and Saunders, 1966).
Here, we characterize two novel peptide toxins from C. californicus, cal12a and cal12b, that block voltage-gated Na+ channels in squid neurons. These peptides have an unusual type 12 Cys-framework (Brown et al., 2005), and molecular cloning and mass spectrometry (MS) yielded their complete sequences and PTMs. At least 15 homologues of these toxins were found by molecular cloning, and constitute the Cal12.1 family of cDNAs. Individual snails were found to express distinct Cal12.1 mRNA variants. Although these peptides appear to be affiliated with the O-superfamily, they represent a new type of conotoxin that blocks Na+ channels. How these toxin isoforms relate to specific targets in the multitude of prey species attacked by C. californicus remains an intriguing question.
MATERIALS AND METHODS
Throughout this report we have used the three-letter designation for Conus californicus toxins (Cal for cDNAs and cal for peptides) to avoid ambiguity with several other species (e.g. Conus canonicus). We have also followed the nomenclature scheme outlined in the ConoServer database (http://research1t.imb.uq.edu.au/conoserver/) as far as practicality allows.
Animals and venom samples
Specimens of C. californicus (shell length 1.5–3.5 cm) were collected from shallow subtidal areas in southern Monterey Bay and La Jolla, CA, USA. Intertidal specimens were also collected in Bahia Asuncion, BCS, Mexico. Adult Loligo opalescens Berry 1911 (Doryteuthis) (Anderson, 2000), Octopus rubescens Berry 1953 and Aplysia californica Cooper 1863 were collected from Monterey Bay. Strombus luhuanus Linn. 1758 was collected in American Samoa and shipped to Hopkins Marine Station, Pacific Grove, CA, USA. Juvenile specimens of Sepia officinalis Linn. 1758 were obtained from the National Resource Center for Cephalopods, Galveston, TX, USA.
Total venom for physiological experiments (Monterey specimens) was extruded from an excised venom duct onto a small piece of plastic film and suspended in 0.5 ml of external recording solution (see below), manually homogenized, heated to 90°C for 5 min and centrifuged at 12,000 g for 15 min. Heating was necessary to avoid lysis of cells during bioassays. The supernatant was then decanted and stored at 4°C. ‘Milked’ venom was also collected by arousing a snail with a small piece of squid (L. opalescens) skin (internal layer) stretched over a 0.5 ml microfuge tube and enticing a venom injection into the vial (Jakubowski et al., 2005). The sample (5–10 μl) was then frozen at –80°C.
Venom samples from up to 30 venom ducts of Monterey snails were prepared for purification (and molecular cloning). For purification purposes, the extruded crude venom was separated into liquid chromatography (LC) fractions as described below and then dried. Individual LC fractions were resuspended in external recording solution (see below) with 0.1% bovine serum albumin for physiological assays. After active fractions were identified, further chromatographic separations were coupled to MS techniques to isolate and identify relevant toxins. For molecular cloning work, venom ducts (after venom extrusion) were frozen in liquid N2 and then used for RNA extraction and reverse transcription (RT) polymerase chain reaction (PCR), as described below. Methodological improvements during the course of the project enabled later biochemical and molecular procedures to be carried out on smaller volumes of venom collected from individual snails.
Chemicals
All organic solvents used in MS were of high performance liquid chromatography (HPLC) grade or better (Thermo Fisher Scientific, Waltham, MA, USA). HPLC peptide standards and all other chemicals were from Sigma-Aldrich (St Louis, MO, USA). Gibco L-15 tissue culture medium was from Invitrogen (Carlsbad, CA, USA).
Original peptide purification and N-terminal sequencing
The contents of 10 venom ducts from Monterey snails were pooled and suspended in 0.1% trifluoroacetic acid (TFA) plus 5% acetonitrile (ACN) in distilled water, manually homogenized and centrifuged as described above. The supernatant was collected, the pellet was re-suspended in 25% ACN plus 0.1% TFA, and the extraction, centrifugation and supernatant removal was repeated. This cycle was repeated with sequentially higher concentrations of ACN (50, 75 and 99.9%), and all supernatant was pooled in a final volume of 1.0 ml.
Venom stocks were fractionated and re-purified by reverse-phase HPLC using a Vydac C18 HPLC column (4.6×250 mm, 5 mm), pre-equilibrated with 5% ACN/0.1% v/v TFA-H2O. Individual peptides were eluted with a standard 1% min–1 gradient of solvent B (solvent A: 0.1% v/v TFA-H2O, solvent B: 90/10% v/v ACN/0.08% TFA-H2O) over 80 min. Samples were eluted at 1 ml min–1 with elution monitored at 214 nm. Seven fractions were collected manually and used for bioassays as described in ‘Electrophysiology’ below.
High levels of Na+ channel-blocking activity were found in a fraction containing at least nine different peptides. A second-stage purification was carried out using a C18 narrow-bore column (2.0 mm) with a 1% gradient of solvent B and a flow rate of 250 μl min–1 and monitored at 210 nm. This yielded a fraction with biological activity that produced a single chromatographic peak.
The remainder of this purified chromatographic fraction was subjected to Edman degradation, N-terminal sequencing (PAN Facility, Stanford University, CA, USA), which unambiguously identified the first 20 amino acids (with the exception of position 17) as DVCDSLVGGHCIHNGC_CDQ (Fig. 1). As discussed in the Results, this sequence characterizes the peptides cal12a and cal12b, with position 17 being a modified tryptophan residue. A low signal-to-noise ratio prevented clear identification of additional residues.
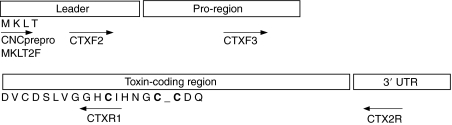
Fig. 1.
Generic structure of the predicted peptide encoded by the Cal12.1.1a cDNA, scheme of the PCR-based cloning strategy and partial amino acid sequence for the mature toxin-coding region as revealed by N-terminal Edman sequencing of the purified peptide. The overall structure of the cDNA is indicated by the boxes. Names, directions and approximate locations of PCR primers used for identifying Cal12.1 variants are indicated. Bold type indicates conserved cysteines.
Later purifications for mass spectrometry
Venom from 30 ducts (Monterey snails) was pooled and homogenized in acidified acetone (40:6:1, acetone:water:HCl). The acetone was subsequently removed using a SpeedVac (ThermoSavant, Holbrook, NY, USA). Analytical HPLC was carried out in 2002–2004 using a RAININ HPXL (Woburn, MA, USA) solvent delivery system monitoring at 220 nm. Approximately 5% of the sample was injected onto a 4.6 mm Vydac C18 column with a flow rate of 1000 μl min–1. A solvent gradient was used to mimic the earlier separations, and fractions were collected every 3 min. Organic solvents were removed from the LC fractions with the SpeedVac, and samples were stored at 4°C.
Fractions from this first stage separation were again used for electrophysiological assays (see below). Although the approximate time of elution had previously been established for the active fraction of interest, differences in LC parameters employed between systems required another identification of the active peptides. Individual fractions were diluted into an external recording solution, with the final concentration of active peptide being estimated from the calculated UV absorbance, ϵ280=2.25×104, based on the cDNA sequence of Cal12.1.1a (see Molecular biology section below).
An active fraction that was eluted for ∼43 min was found to have a doublet peak. This fraction was subjected to second-stage purification using a MAGIC 2002 (Michrom BioResouces, Auburn, CA, USA) micro-bore HPLC, and absorbance (milliabsorbance units, mAU)was monitored at 220 nm and 280 nm. Fractions were separated on a 1.0 mm Discovery BIO Wide Pore column (Supelco, Bellefonte, PA, USA). The separation used a gradient between two solvents, A and B, with solvent A being 95% ACN, 5% acetic acid + 0.1% TFA and solvent B being 90% ACN, 10% H2O + 0.014% TFA. The solvent gradient was 50 μl min–1 from 20–25% B for the first 5 min, 25–40% B for 60 min, and then ramped up to 80% B to flush the column of any remaining compounds.
A post-column split forced 90% of the eluent to flow to the fraction collector, and the remainder was directly sprayed into an LCQ Deca ion-trap (IT) mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). Fractions were collected in 750 μl screw-cap plastic vials using the FC 203B fraction collector (Gilson, Middletown, WI, USA) for offline MS analysis, as described below.
Mass spectrometry
Online electrospray ionization-ion-trap mass spectrometry
Flow from the microbore HPLC was directly sprayed into the LCQ Deca IT mass spectrometer through the electrospray ionization (ESI) source with a spray voltage of 4.3 kV and nebulizing gas flow of 40 units, facilitating desorption. Capillary temperature was 200°C for the split-flow experiments and 220°C for the non-split liquid chromatography–mass spectrometry (LC–MS). The online data were collected using the ‘triple-play’ acquisition method as described in the manufacturer's software. This consists of an initial scan of the full range from 200–2000 mass-to-charge ratio (m/z), a subsequent data-dependent zoomed scan of the 20 m/z window surrounding a precursor ion to decipher charge states, and a final fragmentation scan of the same precursor.
Offline matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and nano-spray ionization mass spectrometry
Direct analysis of LC fractions was possible with minimal sample preparation. For matrix-assisted laser desorption/ionization (MALDI) analysis, 0.5 μl of sample was placed on a gold target followed by addition of 0.5 μl of 2,5-dihydroxybenzoic acid (DHB) matrix (50 μg μl–1 DHB in 50:50 acetone:water). Data were collected in positive-ion linear mode using a Voyager DE-STR time-of-flight (TOF) mass spectrometer (Applied Biosystems, Framingham, MA, USA). Ions were desorbed with a pulsed nitrogen laser (337 nm) and extracted at 20 kV with a 150 ns delay. In parallel, 2–3 μl of each fraction were added to individual PicoTip glass capillaries (New Objective, Woburn, MA, USA) and loaded into the nano-spray ionization (NSI) source. Nitrogen sheath flow was used minimally and varied from 5–15 units depending on the experiment. A spray voltage of 1.6–2.0 kV achieved the best ionization for the peptides. The narrow m/z zoomed-in scan was used to determine charge state and fragmentation, followed by 20–30% collision energy, which allowed the best quality tandem MS (MS/MS) scans. In some cases a third stage of MS fragmentation was necessary to identify a peptide that contained the γ-carboxyglutamic acid (Gla) residue, confirmed by the characteristic loss of 44 Da from the modified amino acid.
Reduction–alkylation procedure
Peptide fractions from the HPLC were partially dried using the SpeedVac as described above. Fractions were added to 100 μl of 50 mmol l–1 NH4HCO3. Reduction of disulfide bonds was accomplished by adding 2 μl of 200 mmol l–1 dithiothreitol followed by incubation at 40°C for 1 h. Alkylation was then carried out by adding 2 μl of freshly prepared 1.0 mol l–1 iodoacetamide (IAA) in 100 mmol l–1 NH4HCO3. The reaction was allowed to proceed in the dark for 1 h at room temperature, after which the reaction was quenched by adding 10 μl of 200 mmol l–1 dithiothreitol. Purification of reduced and alkylated peptides from the reaction mix was accomplished with PepClean C-18 Spin Column centrifugation devices (Pierce, Rockford, IL, USA) following the manufacturer's procedures.
Single-duct venom liquid chromatography–mass spectrometry Later purifications focused on venom isolated from single ducts (Monterey snails). Venom was manually extruded from the ducts and homogenized in a solution of 95% water, 5% ACN and 0.2% TFA. The common procedure was to add 200 μl of this solution to the venom, sonicate the sample for 10 min, vortex, and then centrifuge the insoluble material into a pellet at 12,000 g for 5 min. This solution was frozen until HPLC analysis. Individual 10-μl aliquots of each venom duct extract were injected onto the MAGIC 2002 microbore system. A flow rate of 50 μl min–1 was used with the same gradient and solvents described in the ‘Later purifications for mass spectrometry’ section (above). This separation targeted the late-eluting species, including those constituting the doublet, as described above. The entire LC flow was directed to the LCQ Deca IT ESI probe for detection and analysis of eluting peptides.
Data analysis
Mass spectra were interpreted manually. Observed masses above 5000 Da are reported as their average masses; others are reported as mono-isotopic masses. (A proton is included because masses are reported as singly charged.) Calculated peptide masses were predicted with both PAWS (http://bioinformatics.genomicsolutions.com) and Protein Prospector (Clauser et al., 1999). The charge state was determined by calculating the isotopic distance (if resolved) and/or multiple charge states being present in the spectrum (i.e. a +4 and +5 peak observed in a single full scan). Once a putative match was made to a cDNA, the fragmentation data were compared to predicted MS/MS ions calculated from Protein Prospector's ‘MS-Product’ function. Owing to the high mass and presence of bromines in some cases, an increasing tolerance was used to match monoisotopic predicted masses to the average masses observed in the spectra.
Electrophysiology
Procedures
Adult squid (Doryteuthis opalescens, formerly Loligo) were collected from southern Monterey Bay, CA, and maintained in a circular tank (2 m×1 m deep) with flow-through seawater at ∼15°C for up to 1 week. Giant-fiber-lobe (GFL) neurons were prepared from excised stellate ganglia following established procedures and maintained in vitro for up to 7 days on glass coverslips at 14°C (Gilly et al., 1990). These cells were used for recording voltage-gated Na+ (Gilly et al., 1990), Ca+ (McFarlane and Gilly, 1996) and K+ currents (Ellis et al., 2001) using whole-cell patch-clamp methods at 12–13°C. Electrodes had a resistance of <0.7 MΩ when filled with internal recording solution, and series resistance compensation was employed to the greatest extent possible.
Sodium currents were measured using an external (bath) solution containing (in mmol l–1): 480 NaCl, 10 CaCl2, 20 MgCl2, 20 MgSO4, 10 Hepes (pH 7.8). In some cases, 20 μmol l–1 GdCl3 was added to eliminate Ca+ currents without affecting Na+ currents (unpublished data). The internal (pipette) solution contained (in mmol l–1): 100 sodium glutamate, 50 NaF, 50 NaCl, 300 tetramethylammonium glutamate, 25 tetraethylammonium chloride, 10 sodium EGTA, 10 Hepes (pH 7.8).
Molecular biology
General
Total RNA was isolated from venom ducts using either RNaqueous (Applied Biosystems/Ambion, Austin, TX, USA) or Perfect RNA, Eukaryotic (Eppendorf, Westbury, NY USA) kits and quantified by UV absorbance at 260 nm. mRNA was prepared and fractionated using the SuperScript Plasmid System with Gateway Technology for cDNA Synthesis and Cloning (Invitrogen) according to the manufacturer's instructions. PCR was carried out using Pfu Turbo or Pfu Ultra DNA polymerases (Stratagene, La Jolla, CA, USA) on a reverse-transcribed template or with Taq DNA polymerase according to manufacturers' instructions. Amplifications were performed using a PTC Thermal Cycler (MJ Research, Waltham, MA, USA) with 1.5–2 mmol l–1 MgCl2, 200 μmol l–1 dNTP and 250 nmol l–1 primers. After an initial denaturation of 7 min at 94°C, the reaction mixture was subjected to 40 cycles of 94°C (10 s) followed by 46°C (30 s) and a change to 72°C at a rate of 0.6°C min–1 followed by 1 min at 72°C. After cycling was complete, an additional extension time of 7 min at 72°C was allowed. Purification of PCR products utilized Wizard Prep (Promega, Madison, WI, USA) or Qiaquick (Qiagen, Valencia, CA, USA) kits. Sequencing was accomplished with Big Dye terminator chemistry (Applied Biosystems, Foster City, CA, USA) utilizing either ABI model 377 or 3100 automatic sequencers.
Identification of cDNAs encoding putative peptide toxins
Conventional, degenerate RT-PCR was employed to obtain a cDNA corresponding to the active peptide first purified. RNA was extracted from pooled venom duct tissue (less the extruded venom) from 12 Monterey snails, and cDNA was synthesized using reverse transcriptase (SuperScript II; Invitrogen). A degenerate, antisense (reverse) primer (CTXR1; Table 1 and Fig. 1) was designed based on the partial peptide sequence derived from Edman degradation as described above, and a sense primer (CNCprepro or MKLT2F) was made to match a highly conserved signal sequence for many Conus peptides of the O-type (Duda and Palumbi, 1999; Woodward et al., 1990). The amplified product was subcloned and sequenced. Exact forward primers (CTXF2 and CTXF3) were then designed based on this information and used in conjunction with oligo(dT) primers to amplify the remainder of the coding and 3′ untranslated region (UTR) of the peptide. This sequence was designated Cal12.1.1a.
Table 1.
PCR primers used in this study
Diversity in the Cal12.1 family was revealed by employing the specific PCR primers described above for the analysis of individual snails from all three collection locations. In most cases RNA was extracted from the venom duct plus its venom content, because this procedure substantially increased the yield of RNA (our unpublished data). RNA from an individual duct was reverse transcribed using either the SuperScript II or AccuScript High Fidelity (Stratagene) enzyme. PCR on first-strand cDNA employed the forward primers CTXF2 or CTXF3 and reverse primer CTX2R (Table 1; Fig. 1). PCR products were cloned into the plasmid pCR4 Blunt-TOPO (Invitrogen) and transformed into competent DH5αT1 cells for subsequent plating on ampicillin medium. Individual colonies were picked, suspended in 15 μl H2O, and directly subjected to PCR using the M13F/R primer pair (Table 1). PCR products were purified and sequenced using the nested T3/T7 primers (Table 1). DNA sequences for all Cal12.1 members have been deposited in GenBank.
Identification of additional putative peptide sequences with a cDNA library
RNA was extracted from pooled venom duct tissue (less venom) from 12 Monterey snails, and 12 μg of RNA was used to purify the mRNA with an oligo(dT) column. A total of 0.12 μg of mRNA was then used for cDNA synthesis using the SuperScript II kit. Products were ligated into pSport1 between the SalI and NotI restriction sites and introduced into Top10 Electrocomp cells (Invitrogen) by electroporation. After growth on ampicillin medium, random colonies were inoculated into standard L-Broth overnight. Plasmid was purified with a conventional mini-prep procedure and sequenced using the T7 primer. This approach yielded the full-length sequence (leader, pro-peptide and toxin-coding region) for Cal12.2a and a large number of other sequences encoding putative conotoxins (unpublished work of C.E., T.A.R., Z.L., N. T. Pierce, J.V.S. and W.F.G.). Additional sequences encoding isoforms of Cal12.2 were also generated from this library, with M13F and M13R (or T7) primers (Table 1). PCR products were purified and sequenced as described above. DNA sequences for all Cal12.2 members have been deposited in GenBank.
RESULTS
Block of Na+ channels by C. californicus venom and purified peptide
Sodium currents (INa) recorded from squid GFL neurons were reversibly blocked by both crude and milked venom isolated from C. californicus (Fig. 2A,B). Only one type of Na+ channel, defined on functional and molecular grounds, appears to be expressed in these neurons (Rosenthal and Gilly, 2003). Inward current that remains at the end of a voltage step (and the inward tail current after the step) flow primarily through Ca+ channels (McFarlane and Gilly, 1996), and these channels are not appreciably affected. Experiments were also carried out on K+ currents, but only small and inconsistent effects of C. californicus venom were observed. At least 90% of the K+ channels under the currents analyzed are of one type that is blocked by TsTX-Kα, a peptide toxin isolated from the scorpion, Tityus serrulatus (Mathes et al., 1997; Ellis et al., 2001), and no consistent effect on these squid K+ channels was thus observed.
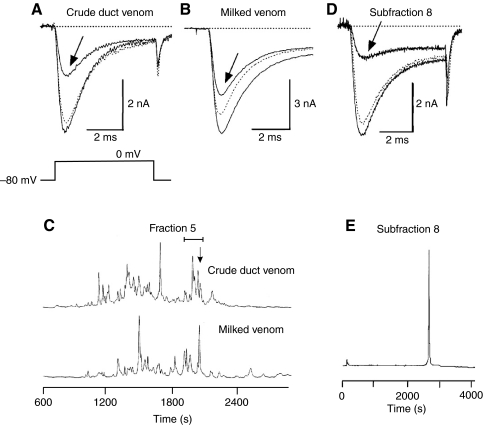
Fig. 2.
Venom components of C. californicus and their effects on voltage-gated Na+ currents (INa) at 0 mV in squid neurons using the whole-cell patch-clamp technique. In A, B and D, the control trace is solid, the test trace (in venom) is indicated by the arrow, and the dotted trace represents the maximum degree of reversal observed after washing out the toxin. (A) Effect of crude duct venom, diluted 1:100 in the external recording solution. (B) Effect of milked venom at a dilution of 1:50 in the external recording solution. (C) HPLC chromatogram of crude and milked venoms. All detectable blocking activity of INa resided in fraction 5. (D) Effect of purified subfraction 8 peptide at a concentration of ∼25 nmol l–1. (E) HPLC chromatogram of subfraction 8.
Active toxin was purified by conventional HPLC from duct and milked venom (Fig. 2C). Purification was carried out only on duct venom, and voltage-clamp assays revealed activity in fraction 5 of the seven tested (bracket in Fig. 2C). A second round of HPLC purification yielded Na+-channel blocking activity in subfraction 8 of nine tested (Fig. 2D), and this subfraction appeared to contain a single peptide species (Fig. 2E).
Voltage-clamp experiments were carried out with neurons from other cephalopods as well as gastropods (data not illustrated). Sodium current was blocked in stellate-ganglion cells of both S. officinalis and O. rubescens by subfraction 8 peptide and raw venom (duct and milked), respectively, at dilutions similar to those found to be effective on squid neurons. No significant effect of duct venom, under comparable conditions, was observed for pedal-ganglion neurons of A. californica, an opisthobranch, or of S. luhuanus, a prosobranch (data not illustrated). Calcium or potassium currents in gastropod neurons tend to be complex because of a mixture of channel types, and these currents were not studied. Of all species tested, only O. rubescens is potentially a natural prey item, and Octopus bimaculatus has been identified in stomach contents of C. californicus (Kohn, 1966).
Primary structure of a novel peptide that blocks squid Na+ channels
Active peptide from subfraction 8 was subjected to conventional N-terminal sequencing, which identified 19 of the first 20 amino acids. Based on this information, a strategy using RT-PCR was devised to obtain the remainder of the amino acid sequence for the N-terminal leader and propeptide region, the toxin-coding region and 3′ UTR (Fig. 3A). The mature toxin-coding peptide begins following an arginine residue in the propeptide region, which is a common basic site for proteolytic processing of Conus peptides (Craig et al., 1999). No 5′ UTR sequence is available.
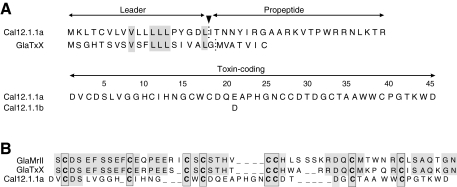
Fig. 3.
Amino acid sequences of Cal12.1.1a and Cal12.1.1b and other framework 12 conopeptides. (A) Full-length amino acid sequences encoded by Cal12.1.1a and Cal12.1.1b cDNAs are compared with the leader and propeptide regions for GlaTxX. Leader and propeptide sequences are identical for Cal12.1.1a and 12.1.1b. Positions of the boundary between the leaders and pro-peptide regions were predicted by SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/). (B) Amino acid alignments of the toxin-coding regions of known peptides with the 8-Cys, type-12 framework. Gaps have been introduced manually to force alignment to the conserved cysteines (in bold) and to maximize amino acid identity in the intra-cysteine loops (shaded areas). Identity in these regions is almost 70% between GlaMrII and GlaTxX, but it is <10% between these sequences and Cal12.1.1a.
This cDNA sequence is designated Cal12.1.1a (GenBank EF644174) in light of the large number of related cDNAs subsequently found (see next section). We have identified the toxin-coding region of Cal12.1.1a in 17 (of 37) individual snails analyzed by RT-PCR (see below) and in 15 clones derived from PCR amplifications of pooled RNA. Amplifications from individual snails also yielded sequences (in ∼25% of the animals) that differed from Cal12.1.1a at only two nucleotides in the toxin-coding region. One substitution (GAA to GAT) leads to a conservative change of Glu to Asp at position 21, but the second substitution (GGT to GGC) is synonymous. This second cDNA is named Cal12.1.1b (GenBank EF644175).
These Cal12.1 cDNAs predict an unusual 8-Cys peptide framework (C-C-C-C-CC-C-C) that has been designated as framework 12 (Brown et al., 2005). This framework clearly differs from that of other conotoxins that target voltage-gated Na+ channels, all of which are 6-Cys peptides of the O- (C-C-CC-C-C) or M-superfamily (CC-C-C-CC) (Terlau and Olivera, 2004). Framework 12 has been reported in only two other cases, GlaMrII and GlaTxX from Conus marmoreus and Conus textile, respectively (Hansson et al., 2004). Alignment of sequences for the toxin-coding regions of these peptides with that of Cal12.1.1a (Fig. 3B) shows that the intra-cysteine loops have essentially no amino acid identity and sizeable differences in length. Sequences of the leader and propeptide regions for GlaMrII are unavailable, but these regions of GlaTxX (Brown et al., 2005) bear little resemblance to those of Cal12.1 (Fig. 3A). Functional properties of the Gla peptides have not been reported.
Identification of two active cal12 peptides and their molecular masses
Further purifications of pooled venom ducts using modified HPLC procedures (see Materials and methods) revealed peptides with reversible Na+-channel-blocking activity eluting between 43–46 min in a doublet peak (region bounded by arrows in Fig. 4A). Peptides eluting in the complex region between 30–37 min also had blocking activity against squid Na+ channels, but these effects were essentially irreversible, and fractions in this region were not studied further.
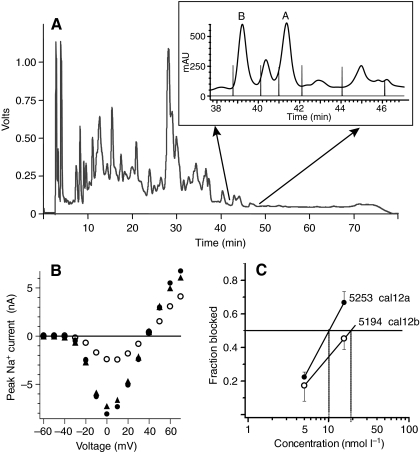
Fig. 4.
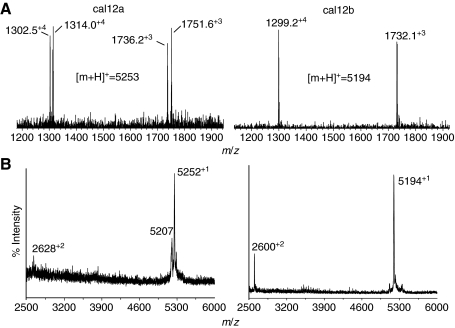
Purification of two active toxins from C. californicus crude venom. (A) HPLC chromatogram of crude extract from pooled venom ducts. A fraction collector was used to isolate the doublet at ∼43 min, which was found to have INa-blocking activity. The inset shows the second-stage separation of material eluting in this region, with vertical lines indicating the collection times for individual fractions. Peaks labeled A and B contained toxins that blocked INa. (B) Effect of the subfraction A on INa in a squid neuron. Filled circles and triangles are control data obtained before toxin application and after washout, respectively; open circles are data obtained in the presence of toxin. (C) Dose dependence of peak A constituent (cal12a, 5253 Da) and that of peak B (cal12b, 5194 Da) in blocking INa at 0 mV in squid neurons. Each point represents mean ± 1 s.e.m. (N=3). Both toxins have an estimated Ki of approximately 15 nmol l–1.
A second-stage chromatographic separation of the active peptides eluting between 43–46 min with a shallow gradient allowed clean separation of two major peaks (peaks A and B in inset to Fig. 4A). Analysis of purified fractions A and B carried out with ESI and MALDI MS revealed molecules with average masses of 5253 Da and 5194 Da (see next section), termed cal12a and cal12b, respectively. Each fraction was individually tested for bioactivity; both peptides reversibly blocked squid Na+ channels at 0 mV with an estimated inhibitor constant (Ki) of 10–20 nmol l–1 (Fig. 4C), with peptide concentrations estimated from relative absorbance. The fraction of INa blocked is approximately twice as large for inward current (at negative voltages) as for outward current (at strongly positive voltages), and this effect was routinely seen with both subfractions.
Mass spectrometry analysis of cal12a and cal12ab peptides and identification with Cal12.1.1 cDNAs
MS analyses of cal12a and cal12b with both IT and MALDI MS revealed that only the peptide of higher molecular mass (5253 Da, cal12a) exhibited a loss of 44 Da (Fig. 5A,B), characteristic of a peptide containing a Gla residue with its extra CO2 moiety (Glu, E to Gla, γ). This PTM commonly occurs in conotoxins (Buczek et al., 2005; Czerwiec et al., 2006; Jakubowski et al., 2006). Examination of the isotopic distribution of the parent mass region of each peptide reveals a mass spectrum containing an ambiguous mono-isotopic peak with a large spread of more than 12 detectable isotopes for both peptides (supplementary material Fig. S1), consistent with the addition of multiple 6-bromotryptophan residues (Trp* or W).
Fig. 5.
MS analysis of active material contained in peaks A and B from the second-stage separation illustrated in Fig. 4. (A) ESI data indicating multiple charge states of the same toxin. The deconvoluted mass based on the centroided data is listed for each toxin. Fraction A (left) contains purified cal12a at 5253 Da that exhibits a 44 Da loss, a feature characteristic of peptides that contain a Gla residue. Fraction B (right) contains purified cal12b at 5194 Da. (B) The corresponding MALDI-TOF data.
Taking these observations into account and considering other common PTMs in conotoxins, particularly hydroxylation of proline residues, we were able to match the cal12a and cal12b peptides to the predicted primary structures for Cal12.1.1a and Cal12.1.1b, respectively (supplementary material Fig. S2). Both peptides contain two hydroxyproline residues, four bromotryptophans, and four nonspecified disulfide bonds. There is only a single Glu residue at position 21 in the primary sequence of Cal12.1.1a corresponding to the Gla residue in cal12a, whereas cal12b has Asp in this position.
Further structural characterization
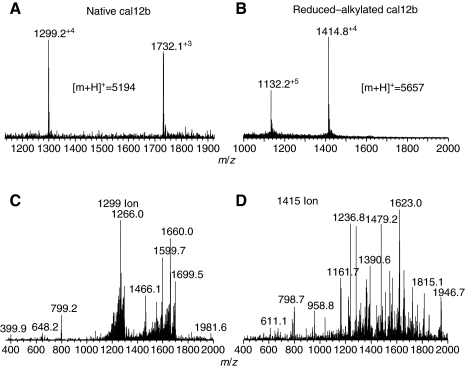
To confirm the number of Cys residues in the mature toxins, we reduced disulfide bonds with dithiothreitol, and alkylated the Cys residues with IAA (Jakubowski and Sweedler, 2004). This resulted in the addition of a carbamidomethyl group to each sulfur atom and a corresponding increase of 58 Da for each Cys residue in the peptide. Both cal12a and cal12b peptides showed an increase of 464±1 Da following this procedure, consistent with eight Cys residues and four disulfide bonds. Fig. 6A,B shows the collected MS data on cal12b before and after reduction and alkylation.
Fig. 6.
MS analysis of disulfide bonds in cal12b. (A,B) The NSI-IT mass spectra collected for the peptide in the native (A) and reduced–alkylated (B) form. The difference of ∼464 Da is due to the eight carbamidomethyl groups (58 Da) on each Cys residue. (C,D) The MS/MS fragmentation spectra of the +4 ions from the corresponding spectra above. Note the increased fragmentation after the disulfide bonds are broken. A more detailed MS/MS analysis of ion 1415 is given in the supplementary material Fig. S3.
Further analysis was performed by fragmenting each peptide before and after the reduction–alkylation procedure. These spectra are shown in Fig. 6C,D. The number of fragments observed was much greater after reduction–alkylation as fragmentation is hindered by intact disulfide bonds. The entire cal12b peptide sequence was characterized using the reduction–alkylation fragmentation spectra. All major peaks in the spectra can be assigned to predicted b or y ions with no peaks over 20% intensity left unmatched (supplementary material Fig. S3). The low-mass fragment ions containing bromine show the characteristic isotopic pattern (supplementary material Fig. S3, inset of y6 ion).
Variability in the presence of cal12a and cal12b peptides in individual snails
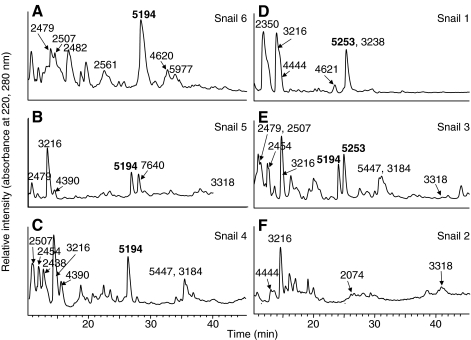
The results above were obtained from pooled venom samples taken from many snails, but we used the same HPLC–MS approach to investigate the presence of cal12a and cal12b isoforms in individual snails, as shown in Fig. 7. Using a chromatographic separation that focused on the late-eluting doublet and other more hydrophobic peptides, we injected ∼5% of the total peptide content extracted from an individual venom duct. In all cases, the resulting mass spectra were complicated with peptides of multiple charge states that co-eluted with the peptides of interest (i.e. 5253=cal12a and 5194=cal12b). Manual interpretation of the IT data revealed parent masses for many of the more abundant species, as indicated in supplementary material Fig. S3, but the identity of these peptides is unknown. Venom from the six animals analyzed contained detectable amounts of either cal12a (Fig. 7A–C) or cal12b (Fig. 7D), both isoforms (Fig. 7E) or neither (Fig. 7F).
Fig. 7.
Toxin expression by individual snails. Peptides were extracted from the venom ducts of six individuals and analyzed by LC–MS, with a focus on the more hydrophobic peptides. cal12a (5253 Da) and cal12b (5194 Da) elute between 25 and 30 min. A snail may express either isoform (A–D), both (E) or neither (F) of these peptides. Some of the more intense parent masses are labeled based on the collected mass spectra from each run, and different snails clearly express different peptides. The peptide at 3216 Da is relatively abundant in five of the six snails, but its identity is unknown.
Although the lack of an observable peak does not prove the absence of any given peptide, the intensity of a peak is related to the relative amount of the peptide present. Thus, there are large variations in the levels of cal12a and cal12b in relation to peak 3216, a prominent peak in five of the six snail extracts. Detection levels for individual peptides are typically ∼100 fmole in the injected sample, and it is possible that peptide levels below this are functionally significant. Nonetheless, these results suggest that substantial variability exists in the levels of cal12a and cal12b in the venom of individual snails.
A family of Cal12.1 cDNAs encoding additional cal12 peptides
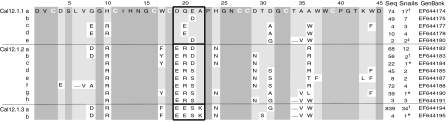
We searched for additional Cal12.1 mRNA variants with the specific amplification primers CTXF2 or CTXF3 with CTX2R (Table 1, Fig. 1) in RT-PCR with total RNA isolates from both pooled samples and from venom ducts of 37 individual snails. Fig. 8 provides an alignment of the toxin-coding regions for 15 Cal12.1 variants that were found in more than one snail, or in separate amplifications from a single snail. Although some regions are absolutely conserved (darker gray areas in Fig. 8, e.g. C11 to C16 and W38 to T42), a high degree of variability exists elsewhere. The pattern of substitution for amino acids 19–22 is particularly interesting and can be used to group the Cal12.1 variants into three subtypes, as indicated by the outlined boxes, in Fig. 8, each of which shows a common amino acid motif in this region.
Fig. 8.
Primary structure of the toxin-coding region of the Cal12.1 variants. Cysteines defining framework 12 and absolutely conserved amino acids are indicated by the darker gray shading. Light gray shading indicates identity with Cal12.1.1a. The region of amino acids 19–22 allows division into subfamilies (Cal12.1–1.3), with each subfamily having a ‘signature’ consensus sequence indicated by the outlined boxes. The number of times each transcript was independently identified in analysis of individual snails is indicated (Seq), along with the number of snails (Snails) in which it was detected with the following notations: †detected more than once in pooled RNA from Monterey snails; *detected in a repeated amplification from the same snail using the same primers; ‡Ca12.1.1e was found in a repeated amplification from a third snail in which it was not initially detected. GenBank accession numbers are given in the rightmost column.
This leads us to propose a nomenclature for members of the Cal12.1.1–3 subtypes by denoting individual members of a subtype by a letter suffix. The peptides cal12a and cal12b correspond to the cDNA sequences Cal12.1.1a and Cal12.1.1b, respectively. This approach allows a practical method of grouping the Cal12.1 members based on a simple structural feature (amino acids 19-22) and can accommodate sequences to be discovered in the future.
Another highly variable region occurs between amino acids 34 and 37, but this region would seem to be less useful in classifying Cal12.1 members. Although the deletion at position 34 in both Cal12.1.3 members is also found in Cal12.1.1e and Cal12.1.2.h, the rest of the sequence in this region would support the Cal12.1.1–3 groupings as proposed.
Amino acid substitutions in both of these variable regions, particularly over amino acids 19–22, are in many cases quite radical in the different subfamilies. For example, an acidic (Glu) or polar (Gln) residue at position 20 occurs in Cal12.1.1 and Cal12.1.3, but a basic residue (Arg) characterizes Cal12.1.2. Similarly, a nonpolar residue (Ala) at position 22 is present in Cal12.1.1 and Cal12.1.2 in contrast to Lys in Cal12.1.3. A similar pattern occurs at position 21, where an acidic residue (Glu or Asp) occurs in Cal12.1.1, but Ser is found in Cal12.1.3. In this case, Cal12.1.2 members show both Asp and Ser at this site.
In contrast to this variability, the flanking regions (spanning amino acid positions 9–27) are highly conserved. Only position 24 shows a structurally significant substitution (His to Asn). The variable region that includes amino acids 34–37 is also adjacent to a highly conserved region (amino acids 38–45). Only a single non-conservative substitution occurs in this region (Lys replaced by Leu at position 43 in Cal12.1.2e). Thus, the most highly variable portions of Cal12.1 isoforms are surrounded by highly conserved regions.
Variability in expression of Cal12.1 transcripts
Examination of cDNAs derived from individual snails revealed a high degree of individuality in the expression of specific Cal12.1 variants. For example, in 12 Monterey snails we found a total of seven different Cal12.1 species, but the number expressed in any given snail was never more than three. Of a total of 422 sequences recovered from these 12 snails, Cal12.1.3a was by far the most common (324 sequences in 11 snails), followed by Cal12.1.2b (56 sequences in 2 snails), Cal12.1.1a (17 sequences in 7 snails), Cal12.1.1b (12 sequences in 2 snails), Cal12.1.1d and Cal12.1.2e (6 sequences in 1 snail each). A single example of Cal12.1.2h was found in one snail.
Snails from La Jolla (11 snails, 371 sequences) and Bahia Asuncion (14 snails, 502 sequences) tended to show even more variability, with nine and 13 different variants being expressed in these locations, respectively. In the most extreme case, two Bahia Asuncion snails were each found to have five different Cal12.1 mRNAs with only Cal12.1.1a and Cal12.1.3a in common. In general, the most commonly expressed sequence overall was Cal12.1.3a, but even here substantial individual variability existed. In four snails it was the only transcript found (19–35 sequences), but in three snails it was not detected at all (22–48 clones sequenced).
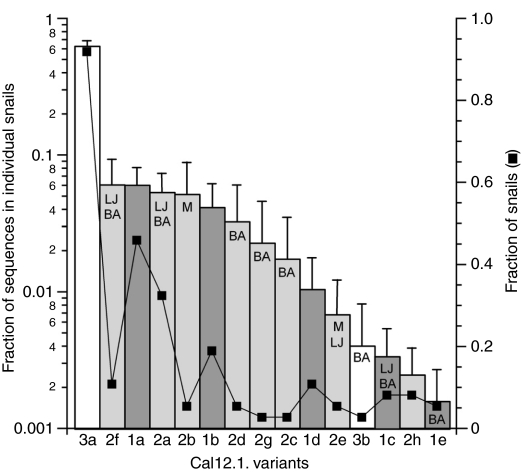
The expression of the Cal12.1 variants by individual snails is summarized in Fig. 9. In general, the more commonly expressed transcripts at the individual level were also expressed in a higher percentage of animals, but considerable variation exists in this relationship. The most abundant species was Cal12.1.3a (63% of total sequences in 92% of the snails) with Cal12.1.1a being the third most frequently expressed transcript.
Fig. 9.
Expression of Cal12.1 transcripts in individual snails. An index of expression for each Cal12.1 variant (indicated on the x-axis) was computed as the mean (±s.e.m.) of fractions found in 37 individual snails. Letters in the bars indicate the locations where each isoform was detected: M, Monterey; LJ, La Jolla; BA, Bahia Asuncion; no label, all three locations. Squares indicate the relative number of snails in which each family member was independently identified. Cal12.1.3a is by far the most commonly expressed transcript; Cal12.1.1a and Cal12.1.1b are expressed at a much lower frequency. The number of sequences independently obtained with RT-PCR analysis from an individual snail varied from 19 to 56 (mean ± s.d.=35±9; total number of sequences=1295). Number of sequences and snails used to compute the plotted values did not include data from amplifications, which were repeated on six snails in the process of confirming various sequences.
Conservation of leader and propeptide regions
Despite the high degree of sequence diversity in the toxin-coding region of Cal12.1 cDNAs, there is virtually no variation in the leader and propeptide regions. Although we have not checked these regions for every member identified, we have done so for the nine of the most frequently expressed variants. This analysis used the RT-PCR primers, MKLT2F and CTX2R, with cDNA from six snails that were selected for their specific individual pattern of expression. As depicted in Fig. 10A, only two sites with amino acid substitutions were identified. Each was the result of a single nucleotide change. Only one example of the Val to Ile change in the leader sequence was found (Cal12.1.2f), but the Asn–Ser variation in the propeptide occurred more frequently (eight Asn, six Ser). In two snails, Cal12.1.2a and Cal12.1.2g showed both Asn and Ser.
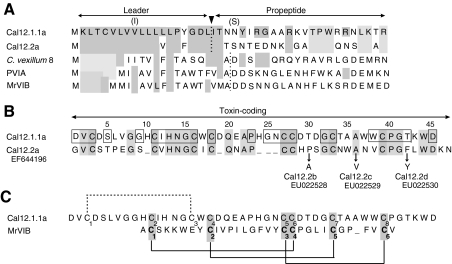
Fig. 10.
Predicted primary structures of Cal12.1.1a and Cal12.2a in comparison with selected O-type toxins from other Conus species. (A) Leader sequences of Cal12.1.1a and Cal12.2a show a high degree of identity, but the propeptide regions are quite different. Identities with Cal12.1.1a are indicated by light gray shading. Alternative amino acids observed in leader and propeptide regions of the Cal12.1 members examined are indicated in parentheses (see text). Leader and propeptide sequences of O-type peptides (C-C-CC-C-C framework) from other Conus species are also shown, and identities with Cal12.1.1a are indicated by the light gray shading. Dotted lines indicate the position of leader sequence cleavage as predicted by SignalP 3.0 Server. Other sequences (see Discussion) are: C. vexillum 8 (Kauferstein et al., 2005), PVIA [C. purpurascens (West et al., 2005)], MrVIB [C. marmoreus (McIntosh et al., 1995)]. (B) Toxin-coding regions of Cal12.1.1a and Cal12.2a. Absolutely conserved regions of the Cal12.1 family are indicated by the boxes. Residues in Cal12.2b–d that differ from Cal12.1.2a are indicated by arrows. GenBank accession numbers are given below the name of each Cal12.2 variant. (C) Toxin-coding regions of Cal12.1.1a and MrVIB, a μO-type toxin. The alignment has been centered on the vicinal cysteines and a single gap introduced in the MrVIB sequence. The known disulfide linkages of MrVIB are shown by solid lines (Daly et al., 2004). If cal12 peptides were linked in this manner, the remaining possibility for the fourth disulfide link is shown by the dotted line. See text for details.
Because the MKLT2F primer includes codons for the start site and the next three amino acids (Table 1), the identity of amino acids 1–4 cannot be rigorously determined. We have never identified a clone for any Cal12.1 member in our cDNA library, so sequences corresponding to these amino acids and the 5′ UTR are not available. As described below, confirmation of the MKLT sequence for the first four amino acids was made in a closely related family of mRNAs.
cDNAs encoding a second type of Cal12 toxin (Cal12.2)
Examination of our cDNA library prepared from pooled venom ducts (Monterey snails) yielded a full-length cDNA that encoded another toxin that shares the 8-Cys-framework of all Cal12.1 members. We have named this sequence Cal12.2a (GenBank EF644196), and its complete amino acid sequence is provided in Fig. 10.
The leader sequences of Cal12.1.1a and Cal12.2a are nearly identical, but the propeptide regions are distinct (Fig. 10A). These leaders show significant amino acid identity with leader sequences of several 6-Cys conotoxins in the O-superfamily, but no similarity exists in the propeptide region.
Comparison of the toxin-coding regions of Cal12.1.a and Cal12.2a (Fig. 10B) clearly indicates that they are related. Some of the absolutely conserved features of Cal12.1 members (boxes in Fig. 10B) are also present in Cal12.2, but little identity exists between Cal12.2a and any of the Cal12.1 members in the distinctive variable regions (amino acids 19–22 and 34–37 of Cal12.1.1a). The gaps at positions 19 and 24–26 in Cal12.2a represent major structural differences from all Cal12.1 members and are likely to have functional consequences.
Three additional Cal12.2 variants were also found in the cDNA library. These sequences are designated Cal12.2b–d, and substitutions in relation to Cal12.2a are indicated in Fig. 10B. Although we have not examined the Cal12.2 toxins as extensively as we have Cal12.1, the small number of variants, two of which are conservative amino acid substitutions, suggest that this group of toxins is much less diverse than Cal12.1. This is consistent with the fact that all three of the Cal12.1 subtypes were identified using a library-based approach analogous to that employed for Cal12.2.
DISCUSSION
The vast majority of previous Conus toxin studies have focused on the rich species diversity of the Indo-Pacific. By contrast, this study examines Conus californicus, which is distantly related to the Indo-Pacific species (Duda and Kohn, 2005; Duda et al., 2001; Espiritu et al., 2001). Because of its high degree of genetic divergence from other Conus species, unique range, and unusually broad diet, we anticipated finding novel peptide toxins in C. californicus. Although we focus here on the only toxin family for which functional effects have been discovered (Cal12.1), we have also identified many other novel sequences that encode putative conotoxins containing 4, 6 and 8 Cys residues (unpublished work of C.E., T.A.R., Z.L., N. T. Pierce, J.V.S. and W.F.G.).
Framework 12 (C-C-C-C-CC-C-C) conotoxins have been previously described (Brown et al., 2005; Hansson et al., 2004), but the relevant peptides (GlaMrII and GlaTxX) bear little similarity to Cal12 cDNAs, other than the predicted framework (Fig. 3). Peptides cal12a and cal12b (corresponding to cDNAs Cal12.1.1a and Cal12.1.1b, respectively) are the first framework 12 conotoxins for which a high-affinity target has been identified. Voltage-gated Na+ channels in squid GFL neurons, thought to be the same as those in giant axons (Rosenthal and Gilly, 2003), are blocked by both toxins with an inhibitor constant (Ki) of ∼15 nmol l–1, with no alteration in the voltage dependence of Na+ conductance or on the kinetics of inactivation.
Peptides in C. californicus venom that block Na+ channels in three species of cephalopods (L. opalescens, O. rubescens, S. officinialis) appear to show no significant activity against Na+ channels in two gastropods (S. luhuanus, A. californica) when tested under the same conditions. We assume that the active peptides in venom preparations tested against both cephalopods and gastropods included cal12a and/or cal12b, and the lack of effect in gastropods suggests that cal12a and cal12b preferentially block neuronal Na+ channels in cephalopods, with gastropod channels being more resistant. Gastropod and cephalopod Na+ channels also show large differences in tetrodotoxin sensitivity and in basic biophysical properties (Gilly et al., 1997). The limited amounts of purified cal12a and cal12b in our study precluded testing gastropod channels at much higher concentrations. Effects on Na+ channels in other taxa, including worms and vertebrates, also remain to be determined.
Given that C. californicus preys heavily on snails (Kohn, 1966), the observed lack of effect of the venom on gastropod Na+ channels seems surprising. Although this may have resulted from the concentration of any active peptides in the venom samples simply being too low to block a significant amount of INa, it may also reflect the fact that neither gastropod tested is a natural prey. Testing gastropods that were actually preyed on by C. californicus would be valuable, as would the testing of Ca2+ and K+ currents in ecologically relevant gastropods.
Assignment of superfamily
Conotoxin superfamilies are defined primarily by their leader sequences (Halai and Craik, 2009; Terlau and Olivera, 2004). A published leader sequence is available for only one other framework 12 peptide, GlaTxX (Brown et al., 2005), but it has no similarity with the leader of the Cal12 members. However, significant identity does exist in the N-terminal leader sequence between Cal12.1 and Cal12.2 members and a number of framework 6/7 (C-C-CC-C-C) conotoxins (Fig. 10A). The MKLTC signal sequence can be considered canonical for the O-superfamily (Duda and Palumbi, 2004), and δ-PVIA from Conus purpurascens and μO-MrVIB from C. marmoreus are O-superfamily toxins (McIntosh et al., 1995; West et al., 2005) that target voltage-gated Na+ channels. The functional effects of the 6-Cys toxin, Conus vexillum 8, are not known, but this sequence also encodes a framework 6/7 conotoxin (Kauferstein et al., 2005).
Homologies in leader sequence clearly associate Cal12 precursor sequences with the O-superfamily, but how does this fit with the striking difference in Cys-frameworks of the toxin coding regions? Conotoxins of the O-superfamily (e.g. MrVIB in Fig. 10C) have a generic structure of C1(3-6)C2(5-10)C3C4(2-8)C5(3-10)C6, where Ci indicates a conserved Cys residue and its position (i=1–6), and the numbers in parentheses indicate the number of intervening amino acids (Gracy et al., 2008). We propose that the basic O-structure is embedded within the 8-Cys-framework defining Cal12 members: C(6-7)C(4)-C1(1)C2(4-8)C3C4(4)C5(4-5)C6, where C1–C6 represent the Cys residues analogous to those of MrVIB. Thus, Cal12 toxins might be considered to consist of a framework 6/7 with two additional Cys residues comprising the N-terminal region of the peptide. Lengths of the inter-Cys segments in the Cal12 sequences fit within the range of the O-superfamily, with the exception of the C1-C2 segment.
We consider this structural similarity in more detail below. This sequence homology and mode of action (block of voltage-gated ion channels) suggest that all Cal12 members should be grouped within the O-superfamily and designated as a new class of conotoxins. We propose ‘O+2’ for this purpose. Inclusion of the only other framework 12 conotoxins (GlaTxX and GlaMrII) in this O+2 classification is questionable because of the extremely different leader sequences (Fig. 3).
We do not know the position of disulfide bonds in cal12 peptides, so it is not possible to evaluate any secondary structural ramifications of the two ‘extra’ N-terminal cysteine residues with respect to other O-superfamily members that target Na+ channels. General three-dimensional structural features appear to be shared between the μO-toxin MrVIB and δ-toxin TxVIA, and in both peptides C1 (adjacent to the N-terminus; see Fig. 10C) is disulfide-bonded to C4, and the intervening region is thought to be important in conferring target specificity (Daly et al., 2004). The analogous positions in cal12 peptides would be C3 and C6 (normal C1–C8 numbering), but the extremely short C3-C4 segment (one amino acid) might interfere with this loop structure. Alternatively, if C2 and C6 of the Cal12.1-encoded peptide were linked as depicted in Fig. 10C, a peptide of secondary structure similar to MrVIB could result. In this case, the fourth disulfide bridge in cal12a would have to be between C1 and C3. Most importantly, the C2-C6 loop would contain the highly variable region between amino acids 19 and 23 that define the Cal12.1.1–3 subtypes. Such a loop could potentially present this segment of charged residues as a reaction surface that is critical to target specificity, similar to the situation thought to exist in MrVIB.
Although Cal12 toxins may be assignable to the O-superfamily, it is difficult to identify a specific functional type. Based on demonstrated functional effects, cal12a and cal12b are not δ-type toxins because they do not affect the inactivation kinetics of squid Na+ channels. A functional relationship to μO-toxins is more difficult to assess. These toxins block both gastropod and vertebrate Na+ channels and also affect Ca2+ channels at high concentration (Daly et al., 2004). Only two closely related μO-toxins (MrVIA and MrVIB) have been identified (Ekberg et al., 2008; McIntosh et al., 1995), and at present there is no compelling reason for assigning cal12a and cal12b to this functional type (Leipold et al., 2007; Zorn et al., 2006).
Despite the hypothesized similarities in secondary structure between cal12a and MrVIB peptides, comparison of primary structural features (sequence, net charge and hydrophobicity) of individual inter-Cys loops of Cal12.1 members with a large number of O-toxins that target Na+ channels (δ- and μO-types) (Heinemann and Leipold, 2007) fails to provide a satisfactory assignment. Although a highly hydrophobic C5-C6 loop (numbering for MrVIB; Fig. 10C) is characteristic of δ- and μO-toxins, as well as Cal12.1 members, low hydrophobicity in the C2-C3 and C4-C5 loops, as exists in Cal12.1, is not typical of δ- and μO-toxins. Given these differences and the uncertainties in secondary structure, we feel that assignment of cal12a and cal12b peptides (and all the other Cal12.1 and Cal12.2 members) to a specific functional type of conotoxins remains an open question.
Posttranslational modifications of cal12a and cal12b
Through MS analysis we have demonstrated that cal12a has a single Gla residue at position 21, whereas cal12b has an Asp substitution at this site. The two other known framework 12 conotoxins contain numerous Gla residues. GlaMrII and GlaTxX contain five Gla residues each, and the modified residues align in the sequences of these peptides (Brown et al., 2005; Hansson et al., 2004). This alignment is not shared by cal12a. Brominated Trp and hydroxylproline residues are also evident in both cal12a and cal12b peptides. Both of these modifications are prevalent in Conus peptides (Craig et al., 1997; Jakubowski et al., 2006; Jimenez et al., 1997; Mann and Jensen, 2003; Steen and Mann, 2002). In most cases, such PTMs cannot be associated with precise functional correlates, but they are thought to increase binding affinity to receptors by altering local peptide charge (Siegfried et al., 1992).
Diversity of the Cal12.1 family
A high degree of diversity clearly exists in the Cal12.1 family of cDNAs, but the 15 different variants reported in this paper represent a conservative estimate. We excluded additional sequences based on two major concerns – potential DNA polymerase errors and recombination of mixed template species during PCR and cloning. A total of 22 sequences were amplified from individual snails (plus four from pooled RNA), but eliminated from further analysis because they were observed only once. We believe that these sequences probably do not represent polymerase errors, for the following reasons. First, we used the more accurate Pfu rather than Taq in the vast majority of amplifications. Second, fidelity of amplification in conjunction with other sequences was extremely high. For example, 363 amplifications from 15 snails designed to search for variants in a putative 6-Cys conotoxin (unpublished work of C.E., T.A.R., Z.L., N. T. Pierce, J.V.S. and W.F.G.) yielded only one hint of a potential polymerase error – a single base change that was seen once in one snail. Nonetheless, we have excluded from this paper any Cal12.1 sequence that was found less than three times in a single snail.
Recombination during PCR or cloning (Meyerhans et al., 1990) is more difficult to assess, and a mixture of Cal12.1 mRNAs in any given snail could theoretically give rise to PCR or cloning-induced chimeras. For example, Cal12.1.1e contains a sequence included in both Cal12.1.1b (amino acids 1–30) and Cal12.1.3a (amino acids 31–45) and could possibly be such a chimera. Similarly, Cal12.1.2h corresponds to the same regions of Cal12.1.2d (or e) and Cal12.1.3a. We therefore deemed a sequence to be valid only if it could not have been a theoretical recombination product of other sequences found in the same snail. This criterion eliminated another four sequences. In the two examples above, only one of the three snails in which Cal12.1.1e was found also showed Cal12.1.1b, and one of the three snails yielding Cal12.1.2h had neither Cal12.1.2d nor Cal12.1.2e. These variants are therefore included in this report.
We feel that some of the sequences that were eliminated in considering both polymerase and recombination errors are likely to be valid. Biological justification for this comes from the fact that of 26 rejected sequences, 18 were found only in snails from Bahia Asuncion. Only six rejected sequences were unique to La Jolla or Monterey (three each). Higher overall diversity in Bahia Asuncion was also evident in the number of reliable Cal12.1 sequences found at the three locations: 13 in Bahia Asuncion (14 snails, 502 total sequences), nine in La Jolla (11 snails, 371 sequences) and seven in Monterey (12 snails, 422 sequences). Moreover, five of the 15 Cal12.1 variants were found only in Bahia Asuncion snails. Monterey snails only showed one unique variant, and La Jolla snails showed none (Fig. 9).
These differences strongly suggest a biogeographical component to expression of novel Cal12.1 transcripts that could potentially be related to dietary differences in the three locations. Although the Monterey and La Jolla snails were collected from subtidal areas, it is likely that potential prey items differ in these locations. The Bahia Asuncion snails inhabited the sand beneath a sea-grass bed during low tide and appeared to forage more widely on an adjacent rocky area at high tide. Based on this extreme habitat difference, these animals probably have a distinct set of prey species along with their unique Cal12.1 toxins. Unfortunately the only gut-content analysis reported for C. californicus was based on La Jolla specimens.
Other types of conotoxins have also been reported to show a high degree of diversity, but in general, variability is considerably less than that observed for Cal12.1 members. To our knowledge, the greatest number (N=18) of reported variants for a single type of toxin in one species was observed for R11 sequences identified in a cDNA library from C. radiatus (Jimenez et al., 2003). Although the pattern of variation is somewhat similar to that described in this paper for amino acids 19–22 of Cal12.1 members, in the case of R11, every Cys-Cys loop showed such variation, with no completely conserved regions. Thus, the variants detected in R11 appear to be more analogous to the differences between Cal12.1 and Cal12.2 members than to the modular differences within the Cal12.1 family. Variation in the Cal12.1 family is clearly different from that in Cal12.2, where it may represent allelic variation (Duda and Palumbi, 2000; Duda and Lee, 2009; Duda et al., 2009) or the diversity among paralogous members of a gene family that have been subject to gene duplication and rapid evolution (Duda and Palumbi, 1999; Conticello et al., 2001; Duda, 2008; Puillandre et al., 2010).
Why so many Cal12.1 variants?
Given the strong structural constraints that are likely to be imparted by the absolutely and/or highly conserved regions of Cal12.1 toxins (i.e. amino acids 9–18, 23–28 and 37–45) and the modular diversity in other regions (amino acids 19–23 and 34–37), it is possible that all isoforms block voltage-gated Na+ channels. This idea is based on a similar modular structure of scorpion toxins that block voltage-gated potassium (Kv1) channels, with selectivity for specific channel isoforms being dictated by amino-acid side chains in variable regions that are flanked by highly conserved sequences (Ellis et al., 2001; Garcia et al., 2001; Gross and MacKinnon, 1996; Miller, 1995). In general, the variable regions form the active surface that defines target specificity.
But why might there be so many Cal12.1 peptides to block only voltage-gated Na+ channels? C. californicus is the most generalist feeder known within the genus, and a correlation thus exists between the diversity of prey types and the battery of putative Cal12.1 peptide isoforms. If they are indeed Na+-channel blockers, many Cal12.1 members probably target molluscs and worms, but some may also be effective in fish. At least six distinct Na+-channel genes exist in teleost fishes, three in molluscs and four in annelid worms (Goldin, 2002), and considerable sequence variation exists between species within each group. Because C. californicus preys on many species within these groups (Kohn, 1966), the existence of such a remarkably large number of Cal12.1 isoforms seems biologically reasonable.
Individuality of expression
That snails of the same species can show such individualistic patterns of expression is a surprising result of this study. Both qualitative and quantitative variations exist in which different Cal12.1 transcripts are expressed in individual snails, and these variations were confirmed at the peptide level for cal12a and cal12b. These findings suggest that the levels of these peptides are at least partially controlled through regulation of gene expression in C. californicus.
Variation in the peptide types in the venom of individuals of another species, C. striatus, has also been reported (Jakubowski et al., 2005). In this case, major qualitative differences between individual snails were limited to milked venom samples, with crude venom in the duct being much more consistent. How this sort of variation in conopeptide types is achieved remains unknown, and it is possible that a similar phenomenon exists in C. californicus.
An additional source of variation in peptide composition should be considered for C. californicus. Although we do not believe that recombination in vitro during PCR amplification or cloning is the source of variability for any of the Cal12.1 variants reported here, naturally occurring recombination could underlie the pattern of variation observed (Espiritu et al., 2001; Olivera et al., 1999). Such recombination would provide another mechanism to accelerate evolution of the toxin-coding regions of Cal12.1 genes, in addition to other mechanisms such as gene duplication.
A specific example discussed above (Cal12.1.1e and Cal12.1.2h) presents an excellent test case to either confirm or refute natural recombination. This inquiry would seemingly require elucidation of the relevant genomic structure of the genes encoding these peptides. To our knowledge, this line of investigation has not yet been carried out for conotoxins from any Conus species. If diversity in the Cal12.1 family does involve recombination in vivo, this particular family must be unusually susceptible to this mechanism, because Cal12.2 sequences show no sign of the modular type of variation evident in the Cal12.1 family.
Clearly, Conus californicus expresses unique toxins. It will be important to determine the full extent of the cal12 peptide families and identify high-affinity targets of other cal12 isoforms and mechanistic details of their actions. It will also be valuable to elucidate the mechanisms responsible for the unusual pattern of variation in the primary structure of these peptides, particularly the potential for natural recombination. Furthermore, this unusual Conus species should shed light on factors that control the highly individualistic pattern of expression of specific Cal12.1 variants and help determine the ecological function of specific toxin isoforms in prey capture.
Supplementary Material
Acknowledgments
We thank Mike Morris, Eddie Kisfaludy, Charles Hanifin and Clayton Gilly for assistance with snail collection and Alex Norton for animal husbandry.
LIST OF ABBREVIATIONS
- CAN
acetonitrile
- DHB
2,5-dihydroxybenzoic acid
- ESI
electrospray ionization
- GFL
giant-fiber-lobe
- HPLC
high performance liquid chromatography
- IAA
iodoacetamide
- IT
ion-trap
- LC
liquid chromatography
- m/z
mass-to-charge ratio
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- MS/MS
tandem MS
- NSI
nano-spray ionization
- PCR
polymerase chain reaction
- PTM
post-translational modification
- RT
reverse transcriptase
- TFA
trifluoroacetic acid
- TOF
time-of-flight
- UTR
untranslated region
Supplementary material available online at http://jeb.biologists.org/cgi/content/full/214/1/147/DC1
The project described was supported the National Science Foundation (NSF) by grant no. IBN-0131788-002 to W.F.G. and by award no. P30DA018310 from the National Institute On Drug Abuse (NIDA) and award no. 5RO1NS031609 from the National Institute of Neurological Disorders and Stroke (NINDS) to J.V.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF, NIDA, NINDS or the National Institutes of Health. Deposited in PMC for release after 12 months.
REFERENCES
- Aguilar M. B., Lopez-Vera E., Ortiz E., Becerril B., Possani L. D., Olivera B. M., Heimer de la Cotera E. P. (2005). A novel conotoxin from Conus delessertii with posttranslationally modified lysine residues. Biochemistry 44, 11130-11136 [DOI] [PubMed] [Google Scholar]
- Anderson F. E. (2000). Phylogenetic relationships among loliginid squids (Cephalopoda: Myopsida) based on analyses of multiple data sets. Zool. J. Linn. Soc. 130, 603-633 [Google Scholar]
- Brown M. A., Begley G. S., Czerwiec E., Stenberg L. M., Jacobs M., Kalume D. E., Roepstorff P., Stenflo J., Furie B. C., Furie B. (2005). Precursors of novel Gla-containing conotoxins contain a carboxy-terminal recognition site that directs gamma-carboxylation. Biochemistry 44, 9150-9159 [DOI] [PubMed] [Google Scholar]
- Buczek O., Bulaj G., Olivera B. M. (2005). Conotoxins and the posttranslational modification of secreted gene products. Cell. Mol. Life Sci. 62, 3067-3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauser K. R., Baker P., Burlingame A. L. (1999). Role of accurate mass measurement (+/– 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871-2882 [DOI] [PubMed] [Google Scholar]
- Conticello S. G., Gilad Y., Avidan N., Ben-Asher E., Levy Z., Fainzilber M. (2001). Mechanisms for evolving hypervariability: the case of conopeptides. Mol. Biol. Evol. 18, 120-131 [DOI] [PubMed] [Google Scholar]
- Cottrell G. A., Twarog B. M. (1972). Proceedings: active factors in the venom duct of Conus californicus. Br. J. Pharmacol. 44, 365P-366P [PMC free article] [PubMed] [Google Scholar]
- Craig A. G., Jimenez E. C., Dykert J., Nielsen D. B., Gulyas J., Abogadie F. C., Porter J., Rivier J. E., Cruz L. J., Olivera B. M., et al. (1997). A novel post-translational modification involving bromination of tryptophan. Identification of the residue, L-6-bromotryptophan, in peptides from Conus imperialis and Conus radiatus venom. J. Biol. Chem. 272, 4689-4698 [DOI] [PubMed] [Google Scholar]
- Craig A. G., Bandyopadhyay P., Olivera B. M. (1999). Post-translationally modified neuropeptides from Conus venoms. Eur. J. Biochem. 264, 271-275 [DOI] [PubMed] [Google Scholar]
- Czerwiec E., Kalume D. E., Roepstorff P., Hambe B., Furie B., Furie B. C., Stenflo J. (2006). Novel gamma-carboxyglutamic acid-containing peptides from the venom of Conus textile. FEBS Lett. 273, 2779-2788 [DOI] [PubMed] [Google Scholar]
- Daly N. L., Ekberg J. A., Thomas L., Adams D. J., Lewis R. J., Craik D. J. (2004). Structures of μO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-senstitive and TTX-resistant sodium channles in mammalian sensory nerurons. J. Biol. Chem. 279, 25774-25782 [DOI] [PubMed] [Google Scholar]
- Duda T. F., Jr (2008). Differentiation of venoms of predatory marine gastropods: divergence of orthologous toxin genes of closely related Conus species with different dietary specializations. J. Mol. Evol. 67, 315-321 [DOI] [PubMed] [Google Scholar]
- Duda T. F., Jr, Kohn A. J. (2005). Species-level phylogeography and evolutionary history of the hyperdiverse marine gastropod genus Conus. Mol. Phylogenet. Evol. 34, 257-272 [DOI] [PubMed] [Google Scholar]
- Duda T. F., Jr, Lee T. (2009). Ecological release and venom evolution of a predatory marine snail at Easter Island. PLoS ONE 4, e5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T. F., Jr, Palumbi S. R. (1999). Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. USA 96, 6820-6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T. F., Jr, Palumbi S. R. (2000). Evolutionary diversification of multigene families: allelic selection of toxins in predatory cone snails. Mol. Biol. Evol. 17, 1286-1293 [DOI] [PubMed] [Google Scholar]
- Duda T. F., Jr, Palumbi S. R. (2004). Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc. Biol. Sci. 271, 1165-1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda T. F., Jr, Kohn A. J., Palumbi S. R. (2001). Origins of diverse feeding ecologies within Conus, a geuns of venomous marine gastropods. Biol. J. Linn. Soc. Lond. 73, 391-409 [Google Scholar]
- Duda T. F., Jr, Chang D., Lewis B. D., Lee T. (2009). Geographic variation in venom allelic composition and diets of the widespread predatory marine gastropod Conus ebraeus. PLoS ONE 4, e6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekberg J., Craik D. J., Adams D. J. (2008). Conotoxin modulation of voltage-gated sodium channels. Int. J. Biochem. Cell Biol. 40, 2363-2368 [DOI] [PubMed] [Google Scholar]
- Elliott E. J., Kehoe J. (1978). Cholinergic receptor in Aplysia neurons: activation by a venom component from the marine snail Conus californicus. Brain Res. 156, 387-390 [DOI] [PubMed] [Google Scholar]
- Elliott E. J., Raftery M. A. (1979). Venom of marine snail Conus californicus: biochemical studies of a cholinomimetic component. Toxicon 17, 259-268 [DOI] [PubMed] [Google Scholar]
- Ellis K. C., Tenenholz T. C., Jerng H., Hayhurst M., Dudlak C. S., Gilly W. F., Blaustein M. P., Weber D. J. (2001). Interaction of a toxin from the scorpion Tityus serrulatus with a cloned K+ channel from squid (sqKv1A). Biochemistry 40, 5942-5953 [DOI] [PubMed] [Google Scholar]
- Espiritu D. J., Watkins M., Dia-Monje V., Cartier G. E., Cruz L. J., Olivera B. M. (2001). Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon 39, 1899-1916 [DOI] [PubMed] [Google Scholar]
- Garcia M. L., Gao Y., McManus O. B., Kaczorowski G. J. (2001). Potassium channels: from scorpion venoms to high-resolution structure. Toxicon 39, 739-748 [DOI] [PubMed] [Google Scholar]
- Gilly W. F., Lucero M. T., Horrigan F. T. (1990). Control of the spatial distribution of sodium channels in giant fiber lobe neurons of the squid. Neuron 5, 663-674 [DOI] [PubMed] [Google Scholar]
- Gilly W. F., Gillette R., McFarlane M. (1997). Fast and slow activation kinetics of voltage-gated sodium channels in molluscan neurons. J. Neurophysiol. 77, 2373-2384 [DOI] [PubMed] [Google Scholar]
- Goldin A. L. (2002). Evolution of voltage-gated Na(+) channels. J. Exp. Biol. 205, 575-584 [DOI] [PubMed] [Google Scholar]
- Gracy J., Le-Nguyen D., Gelly J. C., Kaas Q., Heitz A., Chiche L. (2008). KNOTTIN: the knottin or inhibitor cystine knot scaffold in 2007. Nucleic Acids Res. 36, D314-D319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W. R., Olivera B. M., Cruz L. J. (1988). Peptide toxins from venomous Conus snails. Annu. Rev. Biochem. 57, 665-700 [DOI] [PubMed] [Google Scholar]
- Gross A., MacKinnon R. (1996). Agitoxin footprinting the shaker potassium channel pore. Neuron 16, 399-406 [DOI] [PubMed] [Google Scholar]
- Halai R., Craik D. J. (2009). Conotoxins: natural product drug leads. Nat. Prod. Rep. 26, 526-536 [DOI] [PubMed] [Google Scholar]
- Hansson K., Furie B., Furie B. C., Stenflo J. (2004). Isolation and characterization of three novel Gla-containing Conus marmoreus venom peptides, one with a novel cysteine pattern. Biochem. Biophys. Res. Commun. 319, 1081-1087 [DOI] [PubMed] [Google Scholar]
- Heinemann S. H., Leipold E. (2007). Conotoxins of the O-superfamily affecting voltage-gated sodium channels. Cell. Mol. Life Sci. 64, 1329-1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins C., Grilley M., Miller C., Shon K. J., Cruz L. J., Gray W. R., Dykert J., Rivier J., Yoshikami D., Olivera B. M. (1995). A new family of Conus peptides targeted to the nicotinic acetylcholine receptor. J. Biol. Chem. 270, 22361-22367 [DOI] [PubMed] [Google Scholar]
- Jakubowski J. A., Sweedler J. V. (2004). Sequencing and mass profiling highly modified conotoxins using global reduction/alkylation followed by mass spectrometry. Anal. Chem. 76, 6541-6547 [DOI] [PubMed] [Google Scholar]
- Jakubowski J. A., Kelley W. P., Sweedler J. V., Gilly W. F., Schulz J. R. (2005). Intraspecific variation of venom injected by fish-hunting Conus snails. J. Exp. Biol. 208, 2873-2883 [DOI] [PubMed] [Google Scholar]
- Jakubowski J. A., Kelley W. P., Sweedler J. V. (2006). Screening for post-translational modifications in conotoxins using liquid chromatography/mass spectrometry: an important component of conotoxin discovery. Toxicon 47, 688-699 [DOI] [PubMed] [Google Scholar]
- Jimenez E. C., Craig A. G., Watkins M., Hillyard D. R., Gray W. R., Gulyas J., Rivier J. E., Cruz L. J., Olivera B. M. (1997). Bromocontryphan: post-translational bromination of tryptophan. Biochemistry 36, 989-994 [DOI] [PubMed] [Google Scholar]
- Jimenez E. C., Shetty R. P., Lirazan M., Rivier J., Walker C., Abogadie F. C., Yoshikami D., Cruz L. J., Olivera B. M. (2003). Novel excitatory Conus peptides define a new conotoxin superfamily. J. Neurochem. 85, 610-621 [DOI] [PubMed] [Google Scholar]
- Kauferstein S., Melaun C., Mebs D. (2005). Direct cDNA cloning of novel conopeptide precursors of the O-superfamily. Peptides 26, 361-367 [DOI] [PubMed] [Google Scholar]
- Kohn A. J. (1966). Food specialization in Conus in Hawaii and California. Ecology 47, 1041-1043 [Google Scholar]
- Kohn A. J. (1998). Superfamily Conidae. In Mollusca: The Southern Synthesis. Part B. Fauna of Australia, Vol. 5 (ed. Beesley P. L., Ross G. J. B., Wells A.), pp. 846-854 Melbourne: CSIRO Publishing; [Google Scholar]
- Leipold E., DeBie H., Zorn S., Borges A., Olivera B. M., Terlau H., Heinemann S. H. (2007). muO conotoxins inhibit NaV channels by interfering with their voltage sensors in domain-2. Channels (Austin) 1, 253-262 [DOI] [PubMed] [Google Scholar]
- Luna-Ramirez K. S., Aguilar M. B., Falcon A., Heimer de la Cotera E. P., Olivera B. M., Maillo M. (2007). An O-conotoxin from the vermivorous Conus spurius active on mice and mollusks. Peptides 28, 24-30 [DOI] [PubMed] [Google Scholar]
- Mann M., Jensen O. N. (2003). Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255-261 [DOI] [PubMed] [Google Scholar]
- Mathes C., Rosenthal J. J., Armstrong C. M., Gilly W. F. (1997). Fast inactivation of delayed rectifier K conductance in squid giant axon and its cell bodies. J. Gen. Physiol. 109, 435-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane M. B., Gilly W. F. (1996). Spatial localization of calcium channels in giant fiber lobe neurons of the squid (Loligo opalescens). Proc. Natl. Acad. Sci. USA 93, 5067-5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. M., Jones R. M. (2001). Cone venom – from accidental stings to deliberate injection. Toxicon 39, 1447-1451 [DOI] [PubMed] [Google Scholar]
- McIntosh J. M., Hasson A., Spira M. E., Gray W. R., Li W., Marsh M., Hillyard D. R., Olivera B. M. (1995). A new family of conotoxins that blocks voltage-gated sodium channels. J. Biol. Chem. 270, 16796-16802 [DOI] [PubMed] [Google Scholar]
- Meyerhans A., Vartanian J. P., Wain-Hobson S. (1990). DNA recombination during PCR. Nucleic Acids Res. 18, 1687-1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. (1995). The charybdotoxin family of K+ channel-blocking peptides. Neuron 15, 5-10 [DOI] [PubMed] [Google Scholar]
- Möller C., Rahmankhah S., Lauer-Fields J., Bubis J., Fields G. B., Mari F. (2005). A novel conotoxin framework with a helix-loop-helix (Cs alpha/alpha) fold. Biochemistry 44, 15986-15996 [DOI] [PubMed] [Google Scholar]
- Norton R. S., Olivera B. M. (2006). Conotoxins down under. Toxicon 48, 780-798 [DOI] [PubMed] [Google Scholar]
- Olivera B. M. (1997). E.E. Just Lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Mol. Biol. Cell 8, 2101-2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera B. M., Cruz L. J. (2001). Conotoxins, in retrospect. Toxicon 39, 7-14 [DOI] [PubMed] [Google Scholar]
- Olivera B. M., Walker C., Cartier G. E., Hooper D., Santos A. D., Schoenfeld R., Shetty R., Watkins M., Bandyopadhyay P., Hillyard D. R. (1999). Speciation of cone snails and interspecific hyperdivergence of their venom peptides. Potential evolutionary significance of introns. Ann. NY Acad. Sci. 870, 223-237 [DOI] [PubMed] [Google Scholar]
- Pi C., Liu J., Peng C., Liu Y., Jiang X., Zhao Y., Tang S., Wang L., Dong M., Chen S., et al. (2006). Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics 88, 809-819 [DOI] [PubMed] [Google Scholar]
- Puillandre N., Watkins M., Olivera B. M. (2010). Evolution of Conus peptide genes: duplication and positive selection in the A-superfamily. J. Mol. Evol. 70, 190-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel D., Korn W., Kohn A. J. (1995). Manual of the Living Conidae. Wiesbaden, Germany: Verlag Christa Hemmen; [Google Scholar]
- Rosenthal J. J., Gilly W. F. (2003). Identified ion channels in the squid nervous system. Neurosignals 12, 126-141 [DOI] [PubMed] [Google Scholar]
- Siegfried J. M., Kasprzyk P. G., Treston A. M., Mulshine J. L., Quinn K. A., Cuttitta F. (1992). A mitogenic peptide amide encoded within the E peptide domain of the insulin-like growth factor IB prohormone. Proc. Natl. Acad. Sci. USA 89, 8107-8111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R. J. (1966). Megafauna of the upper miocene castaic formation, Los Angeles county, California. J. Paleontol. 40, 21-40 [Google Scholar]
- Steen H., Mann M. (2002). Analysis of bromotryptophan and hydroxyproline modifications by high-resolution, high-accuracy precursor ion scanning utilizing fragment ions with mass-deficient mass tags. Anal. Chem. 74, 6230-6236 [DOI] [PubMed] [Google Scholar]
- Stewart J., Gilly W. F. (2005). Piscivorous behavior of a temperate cone snail, Conus californicus. Biol. Bull. 209, 146-153 [DOI] [PubMed] [Google Scholar]
- Terlau H., Olivera B. M. (2004). Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol. Rev. 84, 41-68 [DOI] [PubMed] [Google Scholar]
- West P. J., Bulaj G., Yoshikami D. (2005). Effects of delta-conotoxins PVIA and SVIE on sodium channels in the amphibian sympathetic nervous system. J. Neurophysiol. 94, 3916-3924 [DOI] [PubMed] [Google Scholar]
- Whysner J. A., Saunders P. R. (1963). Studies on the venom of the marine snail Conus californicus. Toxicon 1, 113-122 [DOI] [PubMed] [Google Scholar]
- Whysner J. A., Saunders P. R. (1966). Purification of the lethal fraction of the venom of the marine snail Conus californicus. Toxicon 4, 177-181 [DOI] [PubMed] [Google Scholar]
- Woodward S. R., Cruz L. J., Olivera B. M., Hillyard D. R. (1990). Constant and hypervariable regions in conotoxin propeptides. EMBO J. 9, 1015-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn S., Leipold E., Hansel A., Bulaj G., Olivera B. M., Terlau H., Heinemann S. H. (2006). The μO-conotoxin MrVIA inhibits voltage-gated sodium channels by associating with domain-3. FEBS Lett. 580, 1360-1364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.