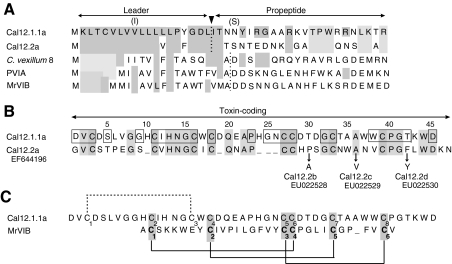
Fig. 10.
Predicted primary structures of Cal12.1.1a and Cal12.2a in comparison with selected O-type toxins from other Conus species. (A) Leader sequences of Cal12.1.1a and Cal12.2a show a high degree of identity, but the propeptide regions are quite different. Identities with Cal12.1.1a are indicated by light gray shading. Alternative amino acids observed in leader and propeptide regions of the Cal12.1 members examined are indicated in parentheses (see text). Leader and propeptide sequences of O-type peptides (C-C-CC-C-C framework) from other Conus species are also shown, and identities with Cal12.1.1a are indicated by the light gray shading. Dotted lines indicate the position of leader sequence cleavage as predicted by SignalP 3.0 Server. Other sequences (see Discussion) are: C. vexillum 8 (Kauferstein et al., 2005), PVIA [C. purpurascens (West et al., 2005)], MrVIB [C. marmoreus (McIntosh et al., 1995)]. (B) Toxin-coding regions of Cal12.1.1a and Cal12.2a. Absolutely conserved regions of the Cal12.1 family are indicated by the boxes. Residues in Cal12.2b–d that differ from Cal12.1.2a are indicated by arrows. GenBank accession numbers are given below the name of each Cal12.2 variant. (C) Toxin-coding regions of Cal12.1.1a and MrVIB, a μO-type toxin. The alignment has been centered on the vicinal cysteines and a single gap introduced in the MrVIB sequence. The known disulfide linkages of MrVIB are shown by solid lines (Daly et al., 2004). If cal12 peptides were linked in this manner, the remaining possibility for the fourth disulfide link is shown by the dotted line. See text for details.