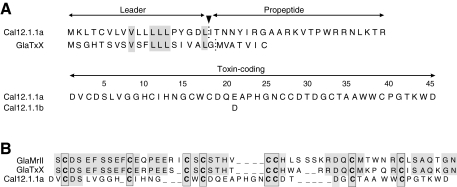
Fig. 3.
Amino acid sequences of Cal12.1.1a and Cal12.1.1b and other framework 12 conopeptides. (A) Full-length amino acid sequences encoded by Cal12.1.1a and Cal12.1.1b cDNAs are compared with the leader and propeptide regions for GlaTxX. Leader and propeptide sequences are identical for Cal12.1.1a and 12.1.1b. Positions of the boundary between the leaders and pro-peptide regions were predicted by SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/). (B) Amino acid alignments of the toxin-coding regions of known peptides with the 8-Cys, type-12 framework. Gaps have been introduced manually to force alignment to the conserved cysteines (in bold) and to maximize amino acid identity in the intra-cysteine loops (shaded areas). Identity in these regions is almost 70% between GlaMrII and GlaTxX, but it is <10% between these sequences and Cal12.1.1a.