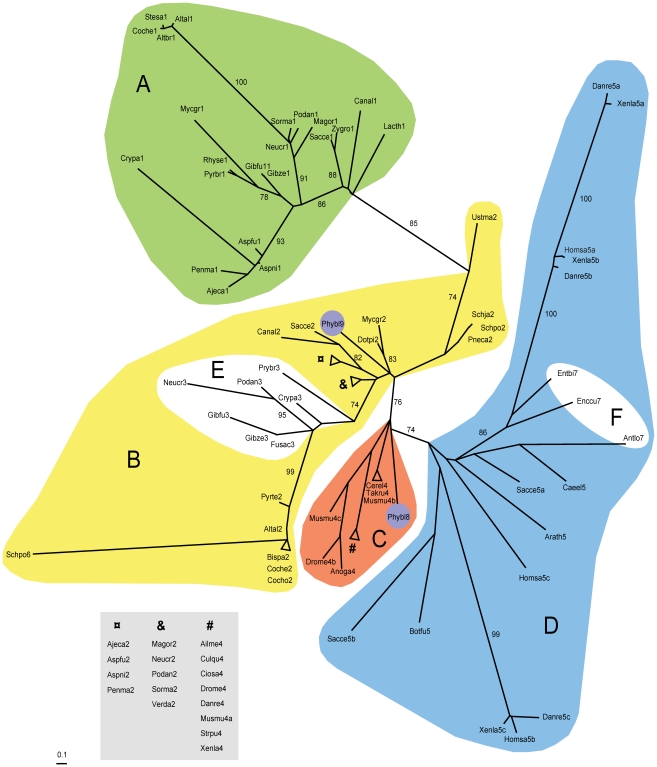
Figure 3. Unrooted phylogram for the HMG superfamily and the α1 domain core amino acid sequences.
Clustering of core amino acid sequences using maximum-likelihood and model LG+G [67]. Labelling is as follows: α1 (A, green), MATA_HMG (B, yellow), SOX (C, orange), HMGB (D, blue), MAT1-1-3 subgroup of MATA_HMG (E, white), Microsporidia MAT sex locus HMG (F, white), Phycomyces blakesleeanus (Zygomycota) sexM (Phybl8) and sexP (Phybl9) are circled in purple. LR-ELW values above 70% are shown. Abbreviations: Ailme, Ailuropoda melanoleuca; Ajeca, Ajellomyces capsulatus; Altal, Alternaria alternata; Altbr, Alternaria brassicicola; Anoga, Anopheles gambiae; Antlo, Antonospora locustae; Arath, Arabidopsis thaliana; Aspfu, Aspergillus fumigatus; Aspni, Aspergillus nidulans; Bipsa, Bipolaris sacchari; Botfu, Botryotinia fuckeliana; Caee, Caenorhabditis elegans; Canal, Candida albicans; Cerel, Cervus elaphus yarkandensis; Ciosa, Ciona savignyi; Coche, Cochliobolus heterostrophus; Crypa, Cryphonectria parasitica; Culqu, Culex quinquefasciatus; Danre, Danio rerio; Dotpi, Dothistroma pini; Drome, Drosophila melanogaster; Enccu, Encephalitozoon cuniculi; Entbi, Enterocytozoon bieneusi; Fusac, Fusarium acaciae-mearnsii; Gibfu, Gibberella fujikuroi; Gibze, Gibberella zeae; Homsa, Homo sapiens; Lacth, Lachancea thermotolerans; Magor, Magnaporthe oryzae; Musmu, Mus musculus; Mycgr, Mycosphaerella graminicola; Neucr, Neurospora crassa; Penma, Penicillium marneffei; Pneca, Pneumocystis carinii; Podan, Podospora anserina; Pyrbr, Pyrenopeziza brassicae; Pyrte, Pyrenophora teres; Rhyse, Rhynchosporium secalis; Sacce, Saccharomyces cerevisiae; Schja, Schizosaccharomyces japonicus; Schpo, Schizosaccharomyces pombe; Sorma, Sordaria macrospora; Stesa, Stemphylium sarciniforme; Strpu, Strongylocentrotus purpuratus; Takru, Takifugu rubripes; Ustma, Ustilago maydis; Verda, Verticillium dahliae; Xenla, Xenopus laevis; Zygro, Zygosaccharomyces rouxii. Numbers after species names indicate α1 proteins (1), MATA_HMG (2), MAT1-1-3 (3), SOX (4), HMGB (5) and other HMG domains (6–9). When more than one domain is present for the same species, the suffix a, b or c was added. Accession numbers of species grouped by evolutionary affinity are in Table S1. Units indicate number of amino acid changes per position.