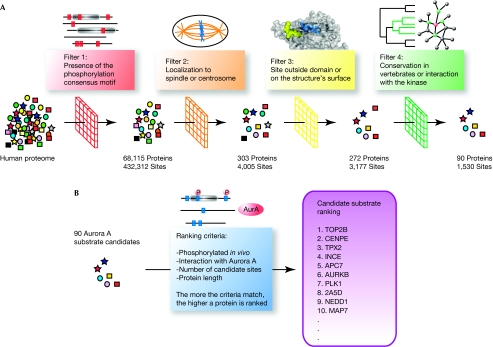
Figure 1.
Schematic representation of the bioinformatics approach. (A) Candidate substrate selection. Aurora substrate candidates were selected on the basis of a series of filters applied to the whole human proteome: presence of an Aurora phosphorylation motif in the sequence, localization to the centrosome or the spindle, accessibility of the consensus motif and conservation of the potential phosphorylation site among vertebrates. (B) Ranking of candidate Aurora substrates (see main text for details).