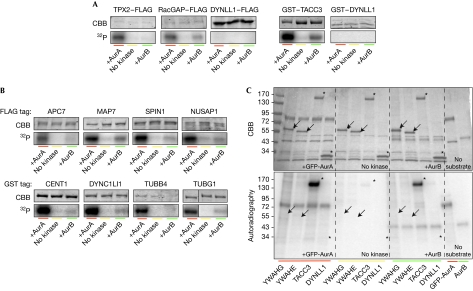
Figure 3.
Validation of candidate substrates by in vitro phosphorylation assays. (A) Specificity of the in vitro Aurora kinase assay tested on known AurA substrates and on a protein not containing the phosphorylation motif (DYNLL1, negative control). Substrates were expressed in human cells, and pulled down by their FLAG or GST tag. Precipitated proteins were mixed with AurA, AurB or only buffer in the presence of 32P-ATP, then separated by SDS–PAGE and stained with Coomassie brilliant blue. Incorporated 32P was visualized by autoradiography. The gel fragments shown belong to representative experiments repeated at least three times. Exposure time of the autoradiographs was adapted for each substrate to allow the visualization of the 32P incorporation. (B) AurA phosphorylation assays were performed as in A on potential substrates from the candidate list. (C) Autoradiography (32P) and Coomassie staining of an in vitro kinase assay gel, where YWHAG (14-3-3γ) and YWHAE (14-3-3ɛ) were tested in comparison with a positive (TACC3) and a negative control (DYNLL1). Arrows and asterisks, positions of candidate proteins. APC7, anaphase-promoting complex 7; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.