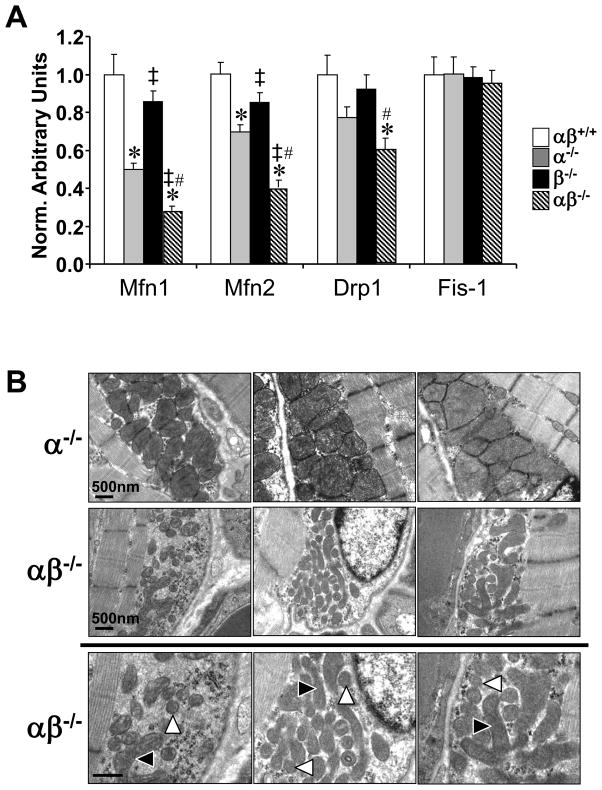
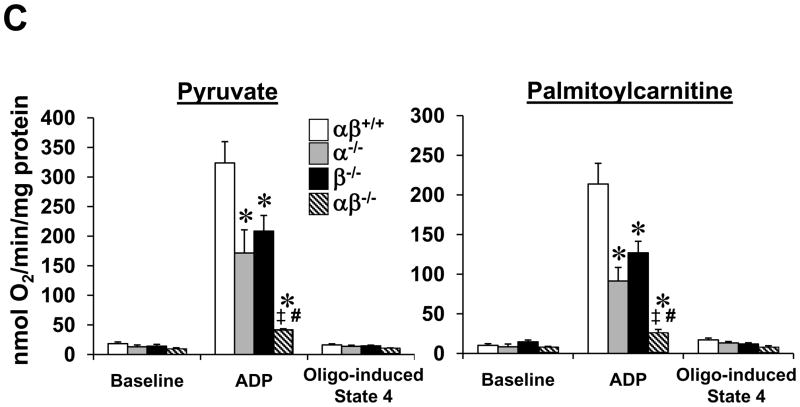
Figure 5. PGC-1α−/−βf/f/MLC-Cre muscle exhibits mitochondrial structural and functional abnormalities and dysregulation of genes involved in mitochondrial dynamics.
(A) Levels of mRNAs encoding mitochondrial fission and fusion genes including mitofusin 1 (Mfn1), mitofusin 2 (Mfn2), dynamin related protein 1 (Drp1 or dmn1l) and mitochondrial outer membrane fission 1 homolog (yeast) (Fis-1) determined by quantitative real-time RT-PCR (n=7–8 per group). Bars represent mean values (+/− SEM) normalized to 36B4 mRNA levels and expressed relative to PGC-1βf/f (αβ+/+) muscle (=1.0). (B) Representative electron micrographs of gastrocnemius showing subsarcolemmal mitochondria in sections from PGC-1α−/−βf/f mice (α−/−; top row) and PGC-1α−/−βf/f/MLC-Cre mice (middle row and magnified in bottom row). Note small, fragmented (white arrowhead) and elongated (black arrowhead) mitochondria. Scale bars represent 500nm. (C) Bars represent mean (+/− SEM) mitochondrial respiration rates determined on mitochondria isolated from the entire hindlimb muscle of the 4 genotypes shown using pyruvate or palmitoylcarnitine as a substrate (4–6 male animals per group). Rates were measured under the following conditions: basal, state 3 (ADP-stimulated), and post-oligomycin treatment (oligo-induced State 4). * p < 0.05 vs. αβ+/+; ‡ p < 0.05 vs. α−/−; # p < 0.05 vs. β−/−. See also Figure S4.