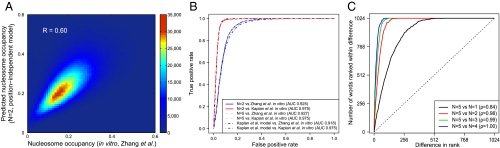
Fig. 1.
Position-independent model predicts in vitro nucleosome occupancy in S. cerevisiae with high accuracy. (A) Density scatter plot for the nucleosome occupancy at each genomic base pair predicted with the N = 2 position-independent model vs. in vitro occupancy observed by Zhang et al. (25). The color of each region represents the number of data points mapped to that region. The model is fit on this data (see Materials and Methods). (B) The receiver operating characteristic curve for discriminating between DNA segments with high and low nucleosome occupancy. The yeast genome was parsed into 500 bp windows and the average nucleosome occupancy was computed for each window. 5,000 windows with the highest and 5,000 with the lowest average occupancies were ranked high-to-low using occupancies predicted with the N = 2 position-independent model, N = 5 position-independent model, and Kaplan et al. model (5). For each partial list of ranked windows with 1,…,10,000 entries we plot the fraction of windows in the list known to have high occupancy on the y-axis, low occupancy on the x-axis. (C) Rank-order plots of energies of 5 bp words: the energy of each word is ranked using position-independent models of order N = 1 through N = 4 and compared with the N = 5 model. Each curve shows the number of words whose ranks are separated by a given distance or less. ρ is the Spearman rank correlation coefficient.