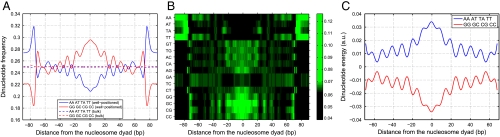
Fig. 2.
Dinucleotide distributions in nucleosome and linker sequences. Nucleosomes were assembled in vitro on the yeast genome using salt dialysis (25). (A) Average relative frequencies of WW (AA, TT, AT, and TA) and SS (CC, GG, CG, and GC) dinucleotides at each position within the nucleosome are plotted with respect to the nucleosome dyad. The relative frequency of each dinucleotide is defined as its frequency at a given position divided by genome-wide frequency. All frequencies are smoothed using a 3 bp moving average. Solid lines: well-positioned nucleosomes defined by five or more sequence reads, dashed lines: bulk nucleosomes defined by one or two sequence reads. (B) Heat map of relative frequencies for each dinucleotide, plotted with respect to the nucleosome dyad. (C) Average energies of WW (AA, TT, AT, and TA) and SS (CC, GG, CG, and GC) dinucleotides at each position within the nucleosome predicted with the N = 2 spatially resolved model are plotted with respect to the nucleosome dyad.