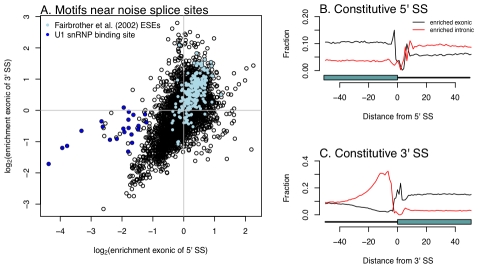
Figure 5. Hexamers enriched near unconserved splice sites are relevant in exon definition.
A. Plotted is the 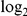 enrichment of all possible hexamers exonic of either 5′ or 3′ noise splice sites. In light blue are hexamers identified as exonic splicing enhancers by Fairbrother et al. [35], and in dark blue are hexamers that are good matches to the consensus U1 snSNP binding site (we include all hexamers matching five contiguous bases of “AGGTAAG”). B and C. Hexamers from A. mark borders of constitutively spliced exons. Each point is the fraction of hexamers starting at that position relative to a constitutively spliced exon (in these cells) which match the hexamers identified as significantly enriched exonic or intronic of the “noise” 5′ or 3′ splice sites.
enrichment of all possible hexamers exonic of either 5′ or 3′ noise splice sites. In light blue are hexamers identified as exonic splicing enhancers by Fairbrother et al. [35], and in dark blue are hexamers that are good matches to the consensus U1 snSNP binding site (we include all hexamers matching five contiguous bases of “AGGTAAG”). B and C. Hexamers from A. mark borders of constitutively spliced exons. Each point is the fraction of hexamers starting at that position relative to a constitutively spliced exon (in these cells) which match the hexamers identified as significantly enriched exonic or intronic of the “noise” 5′ or 3′ splice sites.