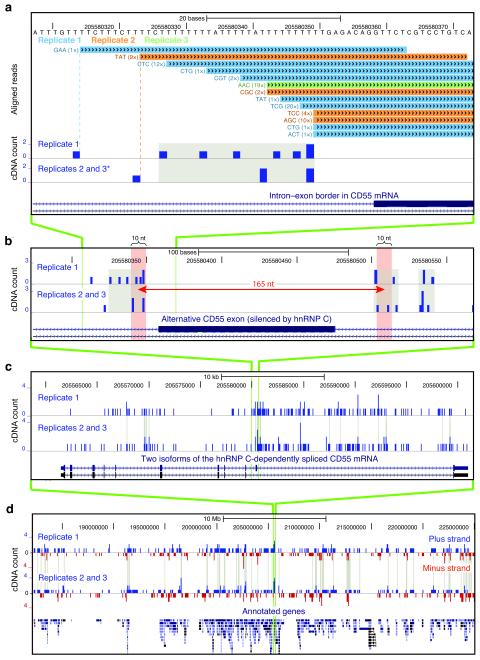
Figure 2.
The genomic location of hnRNP C cross-link nucleotides. (a) Conversion of mapped iCLIP sequence reads into cDNA count values. Genomic sequence is shown above the color-coded positions of cDNA sequences from replicate experiments, preceded by the associated random barcode and the number of sequenced PCR duplicates (given in brackets). In the lower panel, a ‘cDNA count’ was assigned to the upstream ‘cross-link nucleotide’. Cross-link nucleotides within filtered clusters are highlighted in grey. The position of an alternative exon in CD55 mRNA is shown at the bottom. Modified image of the UCSC genome browser (human genome, version hg18, chromosome 1, nucleotides 205,580,308 to 205,580,373). * Due to space limitations, replicates 2 and 3 were merged into one lane. (b) Long-range spaced cross-link nucleotides flank the alternative exon in CD55 pre-mRNA. A distance of 165 nucleotides is marked by a red arrow with red shaded bars on either side representing ten nucleotide surrounding intervals. (c) Cross-link nucleotides are present along the entire length of CD55 pre-mRNA and accumulate around the alternative exon. Clustered cross-link nucleotides are indicated with grey lines. Annotation below shows position of exons in two alternative transcripts. (d) Global view of cross-link nucleotides on chromosome 11 (nucleotides 182,200,000 to 225,000,000). cDNA counts corresponding to positions in plus and minus strand transcripts are shown in blue and red, respectively. Gene annotations are given below. Cross-linking to individual genes and strand specificity are reproduced between replicates.