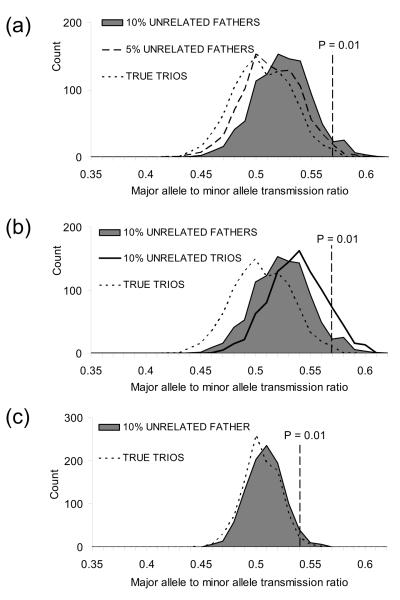
Fig. 1.
Relationship misspecification and over-transmission of common alleles in TDT, simulated with minor allele frequencies of 0.1 in (a) and (b), and 0.4 in (c). The figures investigate the relation between over-transmission bias and: (a) the proportion of pedigrees that are misspecified (5% versus 10% misspecified fathers); (b) the severity of misspecification (misspecified father versus both parents misspecified); (c) allele frequency differences. A bias towards over-transmission of common alleles in TDT in the presence of genotyping error is well-recognized (Heath 1998, Mitchell et al. 2003). The error models considered in previous reports have generally been random errors consistent with imperfect genotyping technologies, rather than the clustered errors one would expect to see as a result of relationship misspecification. Here we demonstrate that relationship misspecification produces a similar bias. Using the SimPed application (Leal et al. 2005), we simulated a neutral SNP marker in a sample of 1000 nuclear family trios, the majority of trios were true parent-offspring trios and no genotyping error was simulated. Each dataset contained a known rate of disguised misspecification (0%, 5% or 10% trios of the 1000 trios) and different misspecification types (Unrelated fathers = trios with an unrelated male disguised as a father; Unrelated trios = three unrelated individuals disguised as a nuclear family trio); and two allele frequencies. Each simulation was repeated 1000 times, and the proportion of major allele transmissions plotted. The transmission ratio equivalent to a TDT p-value of 0.01 is marked on each histogram. When SNP alleles are almost equal in frequency (i.e. a frequency of 0.5), less transmission bias is observed.