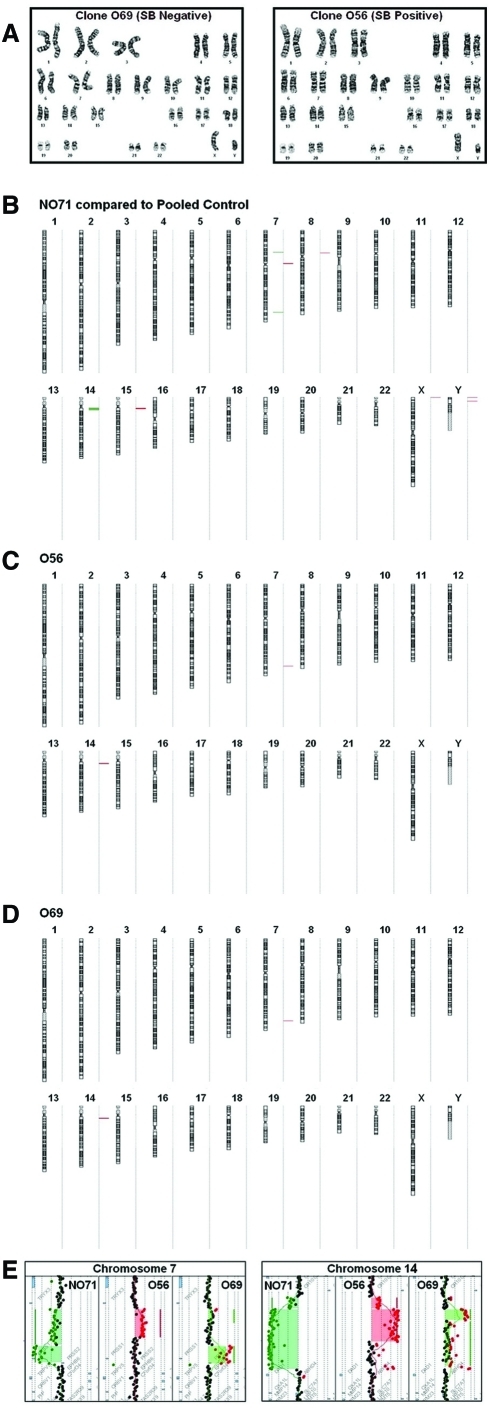
FIG. 6.
Cytogenetic and array CGH analyses of O56 clone. (A) G-band analysis of SB10+ O56 and SB10– O69 clones showing a normal 46,XY male karyotype. (B) Array CGH showing losses at 7q34 and 14q11.2 in the control clone NO71 (compared with pooled male control specimens) and gains in the same regions in clones O56 and O69 (compared with NO71). (C) Array CGH showing copy number gains against control clone NO71 at 7q34 and 14q11.2 regions in clone O56. (D) Array CGH showing copy number gains against NO71 at 7q34 and 14q11.2 regions in clone O69. (E) The control NO71, compared with the pooled control DNA, showed two regions of loss encompassing approximately 346 and 155 kb within 7q34, respectively. The start point for the loss of the 346-kb region was estimated at bp 141663456 and the stop point at bp 142009059. The ratio value of −1 was consistent with a deletion of this region on one chromosome 7 allele. This region contains TCRBV genes. The start point for the loss of the 155-kb region was estimated at 142021348 and the stop point at 142176133. The ratio for this region fell between −2 and −4, consistent with a homozygous loss. Mapped to this region are other TCRB-related genes. Two regions of loss within 14q11.2, encompassing 128 and 633 kb, respectively, were noted. The start point for the 128-kb region was estimated at 21285126 and the stop point at 2142207. Mapped to this region are TCRAV-related genes. The start point for the 633-kb region was estimated at 21420105 and the stop point at 22052858. Mapped to this region are also TCRAV-related genes. Clone O56 shows a ratio of 1.0 for the proximal part of 7q34 and a ratio of 0 for the remaining portion of this 7q34 region. As the control clone NO71 had deletion in these regions, this would indicate that O56 had no loss for the proximal region, and had the same loss as the control NO71 for the distal portion of this region. Clone O69 had a small gain for a portion of the 7q34 region, likely indicating a mixture of cells that had the same loss as the control NO71 and those that had no loss. Clone O69 had a ratio of approximately 2.0 for the distal portion of 7q34, indicating no loss for this region. For the 14q11.2 region, the ratios for O56 and O69 were generated using the control T cell clone NO71, which showed a ratio of −3 for this region. Thus, the +0.3 ratio for O56 indicates that there is no loss for the proximal portion of 14q11.2 region in these cells; the remainder of this region shows a profile consistent with the presence of some cells with loss, and others not. The findings are similar for O69, which shows no loss for the very proximal region, and some cells with loss for the more distal portion of the region. Of note, for NO71 compared with pooled controls, additional gains were seen in the near centromeric regions of chromosomes 7, 8, and 15, and in the pseudoautosomal regions of Xp and Yp. These gains are among the benign copy number variants that are well documented in published databases to occur in healthy controls, and are interpreted as germline variants of no relevance to the present experiments. Color images available online at www.liebertonline.com/hum.