Abstract
TFIIB is the only factor within the multimegadalton transcription complex that is obligatorily required to undergo dissociation and re-association with each round of mRNA transcription. Here we show that a six-amino acid human TFIIB tip region is needed for appropriate levels of serine 5 C-terminal domain phosphorylation and mRNA capping and for retention of the required elongation factor TFIIF. We suggest that the broad functions of this tiny region are used to suppress transcription noise by restricting functional RNA synthesis from non-promoter sites on the genome, which will not contain TFIIB.
Keywords: Gene Expression, Gene Regulation, General Transcription Factors, RNA Polymerase II, RNA Processing, Transcription
Introduction
To induce proper transcription by RNA polymerase II, on the order of 100 polypeptides assemble on DNA, directed by various promoter elements (1). The function of this giant complex is to recruit RNA polymerase to an appropriate site on the chromosome and to activate it so it can accomplish RNA synthesis. Only one of these polypeptides, TFIIB, must be obligatorily released from the template during this process for transcription to proceed. Thus, TFIIB plays a unique role in transcription initiation. Why TFIIB must associate and then be released and what this TFIIB cycle accomplishes is not well understood.
TFIIB is known to both directly recruit RNA polymerase and to alter its enzymatic properties (2–4). The recruitment relies on several protein domains to bind components of the transcription preinitiation complex as well as to bind RNA polymerase. One of these domains is the “B-finger” or “B-reader loop,” which has a tip that approaches the active site of RNA polymerase within the preinitiation complex (5–7). This tip contains a pair of aspartate residues that bind magnesium and are required for full catalysis by RNA polymerase during the initiation phase of transcription (3–4). The assistance presumably provides a quality control function in that RNA polymerases that associate “non-specifically” with the genome, which is without the assistance of TFIIB and the associated preinitiation complex, will have low catalytic activity.
TFIIB has a number of binding partners within the preinitiation complex in addition to RNA polymerase, TATA-binding protein, to direct it to promoter regions, the RNA polymerase elongation partner IIF, and many other factors including acidic activators (8). Despite these many stabilizing interactions, the release of TFIIB is believed to be required during each transcription cycle (9). This occurs during the process of escape when the polymerase is converted into a fully competent RNA synthesis machine.
The physiological purpose of this release is not known but is presumably related to the many changes in the RNA polymerase and the associated machinery that is coupled to the escape process (10–15). These changes set in motion a series of events that allow proper elongation of transcription and couple elongation to RNA processing (16). Central to this program is the phosphorylation of the C-terminal domain of the RNA polymerase. In humans the largest RNA polymerase II subunit has 52 repeats of a heptapeptide (YSPTSPS), and during escape, serine-5 is phosphorylated by TFIIH (17, 18). This triggers the capping of the 5′ end of the RNA by recruiting capping enzyme and enhancing its activity (19–21). Ser-5 phosphorylation also assists in escape by weakening interactions between RNA polymerase and the mediator protein complex (22). Accessory factors play an important role in the appropriate breakup of the preinitiation complex (13) and in the many changes that prepare the elongating RNA polymerase to produce properly processed full-length RNA. The changes are closely coordinated during the promoter escape process, which ends with the RNA capped, with several factors having been modified and having set in motion an elongation transcription complex that has left certain factors behind and is configured to recruit new factors (23–26).
The timing of promoter escape is centrally related to the release of TFIIB, which is controlled by its tip domain (3). That is, small deletions within the tip do not prevent TFIIB from properly joining the preinitiation transcription complex but do cause TFIIB to be released prematurely. This region of TFIIB is in close proximity (6, 7, 27, 28) to the required elongation factor TFIIF (29), which is also believed to influence the C-terminal domain (CTD)2 phosphorylation program during promoter escape (27). After escape, most factors remain on the template to form a scaffold (9). TFIIF travels with the RNA polymerase during elongation, although it may be subject to initial release during the escape process (9, 13, 30). The scaffold is thought to allow for more rapid reinitiation of transcription as only TFIIB and RNA polymerase/TFIF would be required to re-bind to the template after the giant preinitiation complex is first induced and produces a pioneer round of RNA (31). These data raise the possibility that the TFIIB tip cooperates with TFIIF during promoter escape to allow formation of scaffolds and elongation complexes that produce appropriate levels of properly processed RNAs.
The TFIIB tip region consists of the pair of aspartate residues separated by four amino acids. The length of this region is strictly conserved but not the identity of the four amino acids. There is some uncertainty about the structure of this region, and interconvertible structures may exist (5, 7). Its configuration and placement within the preinitiation complex suggest that it may serve as an RNA placeholder and as the RNA grows during initiation the region is displaced, which contributes to the release of TFIIB, similar to the role of domains within bacterial sigma factor (32, 33).
Prior studies of mutations within this region have shown that the aspartates are essential for transcription in vitro, and the connecting four amino acids contribute to transcription but are not essential (3). In the current work we investigate the properties of these mutants with respect to their roles in events occurring during promoter escape by monitoring CTD phosphorylation, RNA capping, and the release of RNA polymerase and TFIIF into elongation phase. We find that the tiny TFIIB tip region within the giant preinitiation complex is required for all of these processes to occur normally. Thus, a six-amino acid region of a single factor is seen to direct proper initiation via assistance with catalysis and proper promoter escape via release of TFIIB and to have the potential to influence the fate of the RNA produced. The requirement that TFIIB be re-bound during each cycle of transcription initiation may have the purpose of minimizing both the production and functionality of transcripts that might arise from RNA polymerases associated with non-promoter sites along the genome.
EXPERIMENTAL PROCEDURES
Proteins and Creation of Transcription Complexes
TFIIB and its mutants were purified as described (3). Human capping enzyme purification was as follows. The pET-Duet-1 vector carrying the human coding sequence of capping enzyme (Open Biosystems) was transformed into and expressed in BL21 (DE3) cells. Cells were sonicated in sodium phosphate binding buffer (20 mm sodium phosphate, pH 7.8, 100 mm NaCl, 0.05% Triton X-100, 5 mm imidazole, 0.5 mm PMSF, and 0.5 mm DTT), and the supernatant was incubated with nickel-agarose beads for 2 h and then washed 5 times with sodium phosphate wash buffer (20 mm sodium phosphate, pH 7.8, 250 mm NaCl, 0.05% Triton X-100, 10 mm imidazole, 0.5 mm PMSF, and 0.5 mm DTT). The beads were then incubated for 1 h with sodium phosphate elution buffer (20 mm sodium phosphate, pH 7.8, 100 mm NaCl, 250 mm imidazole, 0.5 mm PMSF, and 0.5 mm DTT). The supernatant containing capping enzyme was dialyzed in Buffer D (20 mm Hepes, pH 7.9, 100 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, and 20% glycerol) at 4 °C. The preparation of HeLa cell extracts, immunodepletions of TFIIB, purification of immobilization transcription complexes, and in vitro transcription of the AdE4 promoter were as described (3).
Co-transcriptional Capping Assay
The assay was modified from Mandal et al. (12). Preinitiation complexes isolated by the immobilized pulldown assay were resuspended in 25 μl of Buffer D. 25 μg of human capping enzyme was added. Beads were pulsed with 400 μm ATP, CTP, and UTP and 5 μCi of [α- 32P]GTP. After 30 s of pulsing, the reaction was chased with 1.2 mm cold GTP for 4.5min, then terminated in the same manner as in vitro transcription.
Promoter Escape/CTD Phosphorylation Assay
Preinitiation complexes isolated by the immobilized pulldown assay were resuspended in 50 μl of Buffer D. Beads were incubated with either 100 μm ATP, 100 μm each of ATP and CTP, or all 4 nucleotides at 100 μm. Samples were removed at specific times to separate tubes containing 200 μl of Buffer D. Beads were magnetically pulled down, and the supernatant was removed after 1 min. The beads were immediately saturated with SDS loading dye buffer and boiled for 2 min. The beads were pulled down again, and the supernatant was run on a denaturing SDS-acrylamide gel and Western-blotted against TFIIB (C-18, Santa Cruz Biotechnology), Rap74 (C-18, Santa Cruz Biotechnology), polymerase II (8WG16, Covance), or serine 5 (H14, Covance) and serine 2-phosphorylated CTD (H5, Covance).
Yeast Cultures and Extract
Schizosaccharomyces pombe strains were derived from FY191, -lue1, -ade6, and -ura4. In general, procedures followed those described previously (34). S. pombe TFIIB was cloned into pSLF101. pSLF101 contains a lue2 marker and is used as a tetracycline expression system in FY191. Deletion mutants were created by the Stratagene QuikChange mutagenesis kit. Plasmids were transformed using electroporation, and cells were grown on minimal media supplemented with uracil and adenine but lacking leucine. Cultures were grown to an A595 of 0.8. Cells were harvested and processed to make noodles and whole cell transcription extracts as described (35).
RNA Analysis
Cells were grown in minimal media at 30 °C in the presence of doxycycline (at 0.05 mg/ml) to an A595 of 0.5. Cultures were induced with either water or 1 mm hydrogen peroxide (EMD) for 15 min and spun down at 3500 rpm for 2 min. Cells were washed once and resuspended in RNA extraction buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 10 mm EDTA, pH 8.0). Cells were then subjected to vortexing with acid-washed glass beads (Sigma) for 1 min. The sample was then chilled, vortexed again, and subjected to phenol/chloroform/isoamyl alcohol (EMD) extraction. The aqueous layer was removed and subjected to two cycles of phenol/chloroform/isoamyl alcohol extraction. The nucleic acid was ethanol-precipitated and washed with 70% ethanol, then quantified using spectrometry methods. RT-PCR analysis was performed after DNase I treatment of total RNA samples. A total of 40 μg of RNA was treated with 10 μl of Ambion RNase-free DNase I in a 100-μl reaction at 37 °C for 1 h. The DNase-treated RNA samples were purified by phenol-chloroform extraction and precipitation and resuspended in 20 μl of water. Reverse transcription was performed using AMV reverse transcriptase (New England Biolabs) in a 20-μl reaction for 50 min at 42 °C. The reactions were incubated at 85 °C for 5 min to heat-inactivate the reverse transcriptase and treated with RNase H for 20 min at 37 °C. Seven microliters of reverse transcription reactions were used for PCR amplification using standard procedures but reducing the total number of cycles to prevent saturation of product. Products were separated on agarose gels and stained with ethidium bromide. The oligonucleotides used for reverse transcription and to amplify PCR products were the following: apt1 forward (GACGATAGAATCAATTACC) and apt1 reverse (CCATCAGGTGCTTCATCC), atf1 forward (CGTCTCCCGTCAATACTTCC) and atf1 reverse (CTTTGAGCAAGATCACCGC), ctt1 forward (CTCAAATACCGTCCCTGTTTAC) and ctt1 reverse (GAGCTTCCTTGGAACATATGGG), gpa2 forward (CGATTTTTAATGGATTATCTGA) and gpa2 reverse (CCCGCTTCTTTCAGACTGTGTTG), gpx1 forward (CGACTTGGCTCCTAAGGAC) and gpx1 reverse (CTCTCGATATCGTTTTCGAG), hsr1 forward (CCTGAGGCAATGCCCCTTG) and hsr1 reverse (GGTTGATATTGATGCATCGG), pap1 forward (CCGGACAAACTGAGACGTTG) and pap1 reverse (GCCGCTTCAACATCTCTTTCATC), prr1 forward (GCCGTCTTCGAACGGATCCTCCGAC) and prr1 reverse (GCTGTAAGTCTGCTTGAATGGG), sty1 forward (CGTACACAAATATTCGGTAC) and sty1 reverse (CCATGTTGTGAAACGAC), trr1 forward (GGTTGTTATTATTGGGTC) and trr1 reverse (CCAATTCTTCAAGGTAATG), trx1 forward (CAAGTCTCCGACTCTTCTG) and trx1 reverse, (CGCCTTAATAGAAGCCTC) gpd3 forward (GGTTTCGGTCGTATTGGC) and gpd3 reverse, (GCATCGAAGATGGAGGAG), and apt1 forward (GATAATGGCTCTGGTATG) and apt1 reverse (ACGATCGGCAATACCGGG).
RESULTS
The B-tip Is Needed for the Retention of TFIIF during Elongation
Previously, it was shown that TFIIB tip is important during promoter escape (3). TFIIB is normally released when the RNA reaches ∼15–20 nucleotides in length, but mutants lacking the tip of the B-finger were released when the RNA was much shorter (3). The TFIIB finger closely approaches TFIIF in the active site of the preinitiation transcription complex (36). TFIIF is an essential general transcription factor that is recruited along with RNA polymerase II and travels with it during transcription to provide critical elongation support (37). To learn if the tip has a role in the fate of TFIIF, we assayed TFIIF association with the transcription complex using a slightly modified prior protocol (3) for assaying factor retention by TFIIB and its mutants.
Briefly, the E4 promoter template was immobilized, and human preinitiation complexes were isolated after incubation with activator and HeLa nuclear extract. A full complement of NTPs was then added to allow transcription on the template. At various times the templates were washed lightly to dissociate loosely bound factors, and then the supernatant was discarded to remove any unbound factors. Western blots using antibodies to the large TFIIF subunit Rap74 were then used to assess the extent to which TFIIF remained associated with the transcription complexes on the template.
Wild type TFIIB and two tip deletions (D3 and D7) were used. The two mutants were introduced as described previously (3) by first clearing the nuclear extract with TFIIB antibodies and then adding back purified protein. The D3 mutant (missing three amino acids) has nearly normal transcription activity, and the D7 mutant shows only 30% transcription and releases TFIIB early in the escape process (3). Fig. 1 compares the amount of TFIIF large subunit that remains associated with the templates using the wild type and mutant forms of TFIIB as transcription proceeds.
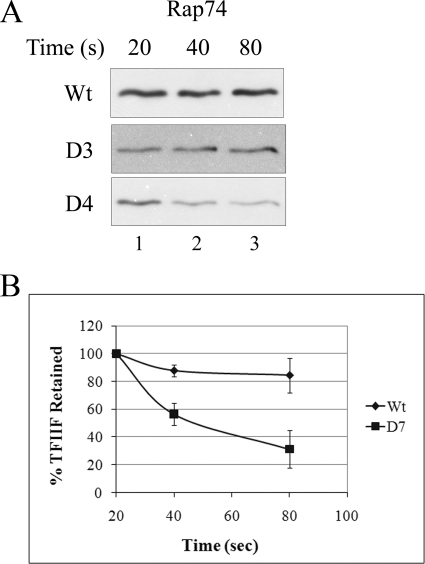
FIGURE 1.
TFIIF retention in elongation complexes. A, recombinant wild type TFIIB containing a full tip region (NDKATKD as residues 57–63) or its D3 (59–61 deleted) and D7 (all 7 deleted) mutants were added to TFIIB-depleted HeLa nuclear extracts, and preinitiation complexes were isolated on an immobilized template. After transcription was begun, samples were processed at the indicated times and probed using antibody against the large subunit of IIF, Rap74. B, quantitative analysis of three independent experiments shows Rap74 retention at the indicated times. Average and S.D. values are shown for TFIIB wild type and D7 at 80 s, respectively: 85 ± 17 and 31 ± 15%.
Fig. 1A, row 1, shows that in a wild type context, TFIIF remains associated with the template throughout, with the level unchanged as transcription proceeds to 80 s. A similar result, no progressive dissociation of TFIIF, was observed with complexes derived from the D3 mutant (Fig. 1, row 2). By contrast, IIF was progressively released from the transcription complexes formed with the larger IIB mutant D7 (Fig. 1A, row 3). By 80 s about 2/3 of TFIIF is released using this mutant (Fig. 1B). Despite this large loss of TFIIF, the starting amount retained by D7 was 80–90% of wild type levels in several experiments, suggesting that D7 complexes bind TFIIF but release it rapidly during initiation.
The result demonstrates that a seven-amino acid region within the TFIIB tip is required for normal retention of TFIIF during transcription. Control experiments using antibodies against RNA polymerase II show that it is not lost from wild type, D3, or D7 complexes (Fig. 2B).
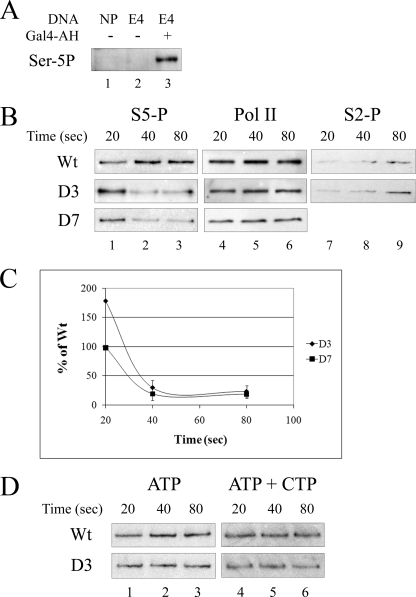
FIGURE 2.
In vitro phosphorylation of RNA polymerase II CTD. Pulled-down templates were assayed for CTD phosphorylation during transcription. A, shown is a demonstration that Ser-5 phosphorylation depends on transcription components by Western blots. The complete system (lane 3) is compared with samples lacking activator protein Gal4-AH (lane 2) or one lacking both template activator sites and a TATA element (lane 1). NP, template with the E4 promoter and activator binding sites removed. B, shown is the effect of TFIIB tip deletion mutants. Samples were removed at the indicated times after transcription had begun and probed with antibody against Serine-5P CTD (lanes 1–3) or RNA polymerase II (Pol II, lanes 4–6) or Ser-2 phosphorylation CTD (lanes 7–9). The wild type, D3, and D7 forms of TFIIB were compared. C, quantitative analysis is shown. Data collected from three independent experiments were quantified and normalized to wild type, whose signal was invariant. The upper curve is the D3 mutant, and the lower is D7. D, shown is an assay before promoter escape. The wild type and D3 mutants were compared as above except that only ATP was added (left column) or only ATP and CTP (right column).
When RNA polymerase is released for transcription elongation upon escape, the remaining general transcription factors stay associated with the promoter to form a transcription reinitiation scaffold in Saccharomyces cerevisiae (9). We assayed for TFIIE to see if this was abnormal using the TFIIB tip mutants. Antibodies to both TFIIE subunits showed that TFIIE remained associated with the template upon transcription using wild type and both mutant forms of TFIIB in human HeLa extract (data not shown).
RNA Polymerase CTD Phosphorylation Is Aberrant with TFIIB Tip Mutations in Vitro
Promoter escape sets in motion a series of timed events that allow proper RNA elongation and processing. The earliest event is the phosphorylation of the CTD of RNA polymerase II on serine 5 of the repeat motif. This is followed by many other critical events, including the capping of the RNA. Eventually Ser-5 phosphorylation diminishes, and Ser-2 phosphorylation occurs (38) as well as other template-associated modifications that support elongation and RNA processing. We used the template immobilization assay previously used to assay CTD phosphorylation in S. cerevisiae (39) to investigate whether the TFIIB tip plays a role in the CTD phosphorylation process. The procedure was essentially identical to that used above to assay factors associated with the template except that antibodies were used to follow phosphorylation of the CTD.
The reliability of the assay in HeLa extracts was assessed. Fig. 2A, lane 3, shows the signal for Ser-5 phosphorylation with appropriate controls in lanes 1 and 2. Reactions that should not produce high levels of transcripts were used as controls. A template without promoter and activator binding sites results in no Ser-5 phosphorylation (Fig. 2A, lane 1). Reactions without activator protein also produce no Ser-5 phosphorylation (Fig. 2A, lane 2). This demonstrates that Ser-5 phosphorylation formation in vitro is dependent on the presence of core promoter and activator. In reactions without NTPs, Ser-5 phosphorylation is also not detectable (data not shown).
Fig. 2B uses this assay to compare the levels of Ser-5 phosphorylation using wild type and mutant forms of TFIIB. This experiment was done as a time course of initial transcription with NTPs being added for 20, 40, and 80 s; these are the earliest times that can be assayed taking account of the time required to isolate the immobilized templates after transcription has begun. The result for wild type (row 1) shows that RNA polymerase remains associated with the template during these times and that Ser-5 CTD phosphorylation can be detected at all three times. In addition, Ser-2 phosphorylation can just begin to be detected at the latest time.
When the D3 and D7 tip mutant forms of TFIIB are used, a different Ser-5 phosphorylation pattern is observed. For D3, Ser-5 is hyperphosphorylated at the earliest time that can be assayed (20 s, Fig. 2B, compare with wild type in column 1). The extent of hyperphosphorylation is nearly 2-fold (see panel C). As transcription proceeds beyond 20 s, Ser-5 underphosphorylation was observed (panel B, columns 2 and 3). These levels are reduced to ∼20% that of the wild type level by 80 s (panel C). For the D7 mutant the hyperphosphorylation at the earliest time was not observed (panel B, column 1), but later underphosphorylation can be seen. The changing Ser-5 phosphorylation levels are not due to loss of RNA polymerase from the template (panel B, columns 4–6). The changes in Ser-5 phosphorylation in the D3 mutant are not accompanied by changes in the levels of Ser-2 phosphorylation (columns 7–9).
The main inference is that the TFIIB tip strongly influences Ser-5 CTD phosphorylation. A larger deletion (D7) that transcribes at only the 30% level (3) allows initial phosphorylation that is prematurely lost. The smaller D3 deletion that transcribes at a nearly wild type level shows rapid hyperphosphorylation followed by the eventual loss that is also associated with D7.
The hyperphosphorylation occurs within 20 s after transcription is initiated by the addition of nucleotides and occurs only with the D7 mutant. To learn if transcription per se is related to this hyperphosphorylation, we restricted RNA synthesis by using limited combination of nucleotides, as described previously in this system (3). The use of ATP provides the phosphorylation substrate and allows open transcription complexes to form but does not support transcription. The use of ATP and CTP allows transcription to begin but was shown to not allow escape. Fig. 2, panel D, shows that none of the changes in phosphorylation patterns seen above is evident when transcription is restricted. We infer that the tip influences Ser-5 phosphorylation but only after transcription has begun. This influence must occur during the escape phase because TFIIB is released before this phase is complete.
The B-tip Influences RNA Capping Levels in Vitro
The phosphorylation of Ser-5 on the RNA polymerase II CTD is required for appropriate RNA 5′ cap formation as the modification recruits and stimulates capping enzyme (39). Capping is thought to occur at the end of the escape process. The above data raise the possibility that capping might be altered, especially with the D3 tip mutant, as this mutant directs hyperphosphorylation during escape and hypophosphorylation at later times.
We adapted an established assay to determine the levels of co-transcriptional capping on these immobilized templates (40). Because capping is inefficient in nuclear extracts, recombinant capping enzyme containing both the triphosphatase and guanylyltransferase was added back to immobilized templates containing transcription preinitiation complexes. [α-32P]GTP along with ATP, CTP, and UTP were added to allow transcription to begin; the radioactive GTP is known to label the 5′-RNA cap in this assay (12) but does not otherwise label the RNA as the template is a G-less cassette. After 30 s, the complexes were chased with nonradioactive GTP for 5 min to suppress artifactual labeling of any transcripts that might exist outside the G-less cassette associated with the promoter. Two parallel assays of RNA were done. The one just described measures the radioactivity associated with the cap of the full-length RNA. In parallel, a separate assay was done to assess RNA levels using radioactive nucleotides incorporated into the RNA body. This serves as an internal control to allow the extent of capping per RNA to be measured. An example of this procedure is shown in Fig. 3.
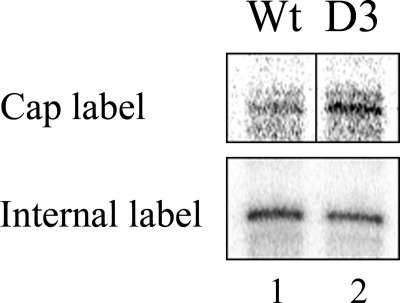
FIGURE 3.
Co-transcriptional capping in vitro. Transcribing pulled-down G-less templates in the presence of capping enzyme were assayed for radioactive ribonucleotide incorporation into the 5′ RNA Cap (lane 1) or the RNA body (lane 2). The body was labeled using ATP, CTP, and [α-32P]UTP for 5 min. The Cap was labeled with ATP, CTP, and UTP with [α-32P]GTP added for 30 s followed by unlabeled GTP for 4.5 min. The wild type and D3 forms of TFIIB are indicated.
The data in row 1 of Fig. 3 shows that the D3 RNA is more heavily labeled with the capping GTP substrate than is the wild type RNA. The parallel transcription assay in row 2 shows that there is somewhat more actual wild type transcript than D3 in this experiment. When normalized, the results of repeated experiments showed that the D3 transcripts were capped at 180% the efficiency of wild type transcripts. This result is likely related to the increased efficiency of Ser-5 phosphorylation of the RNA polymerase CTD using the D3 mutant, as shown above. It appears that the hyperphosphorylation, which occurs during escape, triggers the excess capping, which also occurs during escape. The subsequent premature loss of Ser-5 phosphorylation in the D3 mutant obviously does not block this effect.
Effects of the B-tip Mutations in Vivo
The data have shown that the TFIIB tip alters RNA polymerase Ser-5 CTD phosphorylation. The mutant D3 removes just three amino acids between the catalytic aspartate residues and has profound effects in vitro. We wished to learn if this region of the tip was also important for phosphorylation in an in vivo context. We chose S. pombe as a genetically amenable organism for this purpose. Prior studies (35) have shown that transcription in this organism is much closer than S. cerevisiae to that of mammalian systems. An overexpression system exists in which TFIIB and other general transcription factor mutants have been characterized in S. pombe (34). A study of the role of TFIIB in transcription initiation has suggested that S. pombe TFIIB has properties that better mimic those of human TFIIB (35). In addition, S. pombe extracts containing cloned general transcription factors have been shown to contain a complement of factors that faithfully direct RNA polymerase II transcription (34, 41).
The initial goal of the studies is to learn if deletion of the tip residues leads to changes in RNA polymerase Ser-5 CTD phosphorylation in vivo. The S. pombe TFIIB tip deletion mutant (D4) was created for this purpose. It has four amino acids removed (67EASG70). The four amino acids do not include the two adjacent catalytic aspartate residues (Asp-66 and -71), similar to the human D3 mutant that removes three amino acids between the two aspartates. The human and S. pombe tip sequences are aligned in Fig. 4A. Both wild type and D4 forms of S. pombe TFIIB were transformed into S. pombe FY191, and doxycycline was added to induce overexpression as described (35). The dominant negative system is known to result in overexpression of cloned TFIIB in a background containing the endogenous wild type TFIIB expressed from the chromosome. Effects of mutants of TFIIB and other general transcription factors can be observed in such studies, but they can be muted by the presence of wild type factors (35).
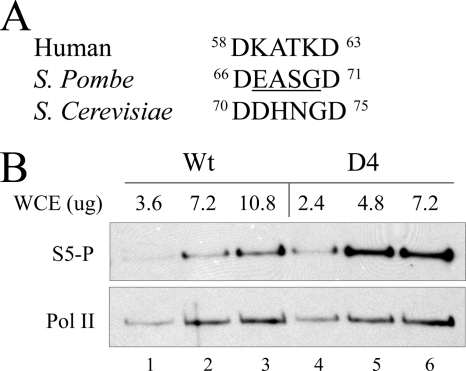
FIGURE 4.
CTD phosphorylation in vivo. A, sequence alignment of human, S. pombe, and S. cerevisiae TFIIB finger motif is shown. B, extracts of S. pombe transformed with either the wild type or D4 form of TFIIB were assayed by Western blots using antibodies against ser-5P (top) or RNA polymerase (bottom). The amount of extract used is shown. Two independent experiments were averaged, showing that the D4 signal was 3-fold greater than wild type. WCE, whole cell extract; Pol II, RNA polymerase II.
To assay the effect of deletion within the TFIIB tip on S. pombe, RNA polymerase CTD phosphorylation standard transcription extracts were made from cells containing wild type or D4 mutant forms of TFIIB. Fig. 4B shows these data in the form of a titration using different amounts of extract. The data show that at each amount of extract the D4 mutant produces a significantly higher level of Ser-5 CTD phosphorylation (row 1). There is no difference in the total amount of RNA polymerase II in the samples between D4 mutant and wild type (row 2). We infer that the deletion of the TFIIB tip can cause higher levels of Ser-5 CTD phosphorylation in vivo as it was capable of doing in vitro.
Next, we wished to determine whether adverse physiological consequences could be attributed to mutants in the TFIIB tip. To do this we screened for growth defects associated with moderate to extreme stresses. Because endogenous wild type TFIIB is present in the constructs, such effects might need to be fairly severe to be detected. The initial screening was for moderately cold temperatures or elevated salt levels and for oxidation stress. These experiments used the D4 mutant and also the related mutant D3, in which Glu-67 was retained. The preliminary results showed that only oxidation stress gave a detectable growth advantage to wild type over mutant TFIIB.
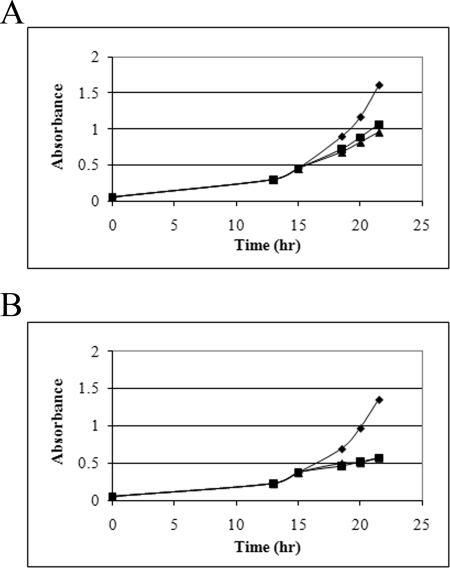
Data showing the difficulty of adjusting to oxidation stress with the TFIIB tip mutation are shown in Fig. 5. S. pombe constructs containing either wild type, D3, or D4 forms of TFIIB were induced for overexpression and then grown to an A595 of 0.5. Peroxide was then added, and growth was followed subsequently by taking absorbance measurements. Cells to which peroxide was not added were followed in parallel (diamonds in Fig. 5). For the wild type TFIIB, the addition of peroxide led to detectably slower growth as the cells adapted to the oxidation challenge (panel A, lower curves were with peroxide). With the D4 mutant, the cells nearly ceased growing when peroxide was added (panel B, lower curves were with peroxide). The D3 mutant was also studied this way and showed a slightly less extensive defect in adjustment to stress (not shown). The defects caused by the two mutations continued beyond the 25-h time shown, with the wild type strain continuing to grow in contrast to the mutants, in which growth is nearly halted (not shown). We infer that mutants in the TFIIB tip can cause difficulty in adjustment to redox challenge in S. pombe.
FIGURE 5.
Oxidation stress adaptation in S. pombe. Cells overexpressing either wild type or D4 TFIIB were challenged with either 1 mm hydrogen peroxide (squares) or 2 mm (triangles) or not challenged (diamonds), and growth was measured at the indicated times. A, wild type TFIIB; B, D4 mutant.
To determine whether RNA imbalances are associated with this poor response to redox challenge, we investigated RNAs levels of genes involved in the redox response. 11 genes have been identified that are at least 2-fold induced by this stress (42–44). The RNA levels from these genes were assessed after cells grown to A595 0.5 were challenged with hydrogen peroxide and then grown for 15 min. Isolated RNA was reverse-transcribed into cDNA using specific primers at the 3′ end of each of these 11 genes. These were then amplified by PCR using facing 5′ primers for a small number of cycles to allow semiquantitative analysis. The data were compared using strains containing wild type TFIIB and the D4 tip mutant. This experiment was repeated three times, and the data from the six different RNA preparations were analyzed.
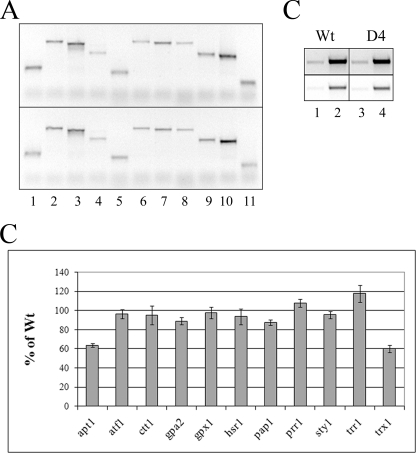
The results of this analysis (Fig. 6) showed that 9 of the 11 genes do not exhibit differences in RNA levels when mutant and wild type are compared but that 2 genes do show a difference. The apt1 and trx1 RNA levels are 40% reduced (see the outer bars with error estimations in Fig. 6) in the D4 mutant strain compared with the wild type strain. Trx1 codes for a thioredoxin, which has a clear relation to redox challenge, but the role of the adenine salvage gene apt1 in stress is unclear. Because both wild type and mutant strains also contain wild type TFIIB from the chromosome, the observed reductions are likely to be underestimates of the effect of TFIIB mutation, and effects on other genes assayed could be hidden. We infer that mutations in the TFIIB tip, which induce aberrations in RNA polymerase phosphorylation and capping of RNA in vitro, can also cause imbalances in RNA levels in vivo.
FIGURE 6.
RNA levels of peroxide-inducible genes. RNAs from S. pombe cells overexpressing either wild type or D4 forms of TFIIB were reverse-transcribed, amplified by PCR, and viewed on agarose gels. A, the top panel refers to wild type, and the bottom panel refers to D4. Genes assayed were: lane 1, apt1; lane 2, atf1; lane 3, ctt1; lane 4, gpa2; lane 5, gpx1, lane 6, hsr1; lane 7, pap1; lane 8, prr1; lane 9, sty1; lane 10, trr1; lane 11, trx1. B, data are from three independent experiments. apt1 was reduced to 64 ± 2, and trx1 was reduced to 60 ± 4% in D4 compared with wild type. C, RNAs from non-inducible genes, gpd3 (lanes 1 and 3) and act1 (lanes 2 and 4), were used as controls. Top row is higher exposure of bottom row.
DISCUSSION
RNA polymerase II transcription is thought to use on the order of 100 polypeptides, but only one of these, TFIIB, undergoes an obligatory association/dissociation cycle with each round of transcription. During each round TFIIB inserts its tip domain within the active site of RNA polymerase and activates it. In this report we showed that the tip controls processes occurring during promoter escape that are centrally linked to the elongation of phases of transcription. We monitored three of the many relevant changes, RNA polymerase II CTD phosphorylation, RNA capping, and TFIIF retention, all of which are abnormal when the TFIIB tip is mutated. These and other processes are essential to complete the factor recruitment and exchange programs during transcription that modify chromatin and allow the mature mRNA to form after multiple processing events (11, 45). These roles are in addition to the previously established role of the tip in transcription initiation (2–3, 46). The TFIIB tip does not play an essential role in either the recruitment of TFIIB or RNA polymerase (3). Thus, a six-amino acid region in a multimegadalton transcription complex is specifically designed to modify the properties of RNA polymerase II such that it can initiate, elongate, and properly process mRNA transcripts.
Why is RNA polymerase II transcription designed to rely on this domain? One likely possibility is that the tip functions to minimize transcription noise, a very serious issue in the genome. Recent studies (47) have suggested that much of the genome can be transcribed into RNA, albeit at low levels, although there have not been suggestions that most of this RNA is functional. The need to suppress transcription noise is evident from the ability of RNA polymerase II to bind avidly to damaged DNA and to transcribe from such sites and indeed from many non-promoter sites in vitro (48). Estimates of steady state levels of damaged DNA that can potentially attract RNA polymerase include nicks every 50 kb (49) as well as other abundant damage sites such as those caused by depurination (50). The total of these nonspecific RNA polymerase II entry sites likely approaches 100,000 sites per genome, far in excess of estimates of active RNA polymerase II promoter sites (51). RNA polymerase II at these sites should not contain TFIIB, as general transcription factors are not required for binding and initiating from nicked DNA.
The absence of TFIIB and its tip at these sites should have several beneficial consequences in reducing transcription noise. As shown previously, the lack of catalytic aspartates will reduce transcription initiation efficiency (3). The data shown here suggest that the RNAs that are produced will be associated with inappropriate elongation complexes that may have reduced functionality. The instability of TFIIF association should make it difficult for such transcripts to be elongated properly. Other aberrations occurring during promoter escape associated with the monitored changes in CTD phosphorylation and mRNA capping indicate that those complexes that manage to initiate and elongate will be in appropriately modified during downstream RNA processing events. Thus, the large size of eukaryotic genomes may require that RNA polymerase II be a deficient core enzyme in the sense that it requires the TFIIB tip each time it makes a proper mRNA. The modest cost of this would be more than balanced by the benefit of noise reduction from non-promoter sites in the genome.
It is not known exactly how the TFIIB tip region contributes to these multiple functions. The data above show that this is largely accomplished after initiation, during the escape process. The tip is known to delay the release TFIIB until the appropriate point during escape (3). TFIIB might function partly via TFIIF in some of these events. TFIIF approaches TFIIB in the active site of the preinitiation complex, and there appear to be mutual effects on function (6–7, 28, 36, 52). TFIIF is a required elongation factor, and prior studies using purified human factors and in yeast extracts show that it can be unstably associated with the preinitiation complex (9, 13, 30). Our data (Fig. 1A) show that TFIIF is not released upon transcription initiation in the context of the human activated preinitiation complex, consistent with its role as an essential elongation factor (53). Small deletions in the TFIIB tip support retention of TFIIF, but the larger seven-amino acid deletion leads to inappropriate release of TFIIF upon transcription. This deletion also leads to the early release of TFIIB, which data show precedes the release of TFIIF (3). Thus, it is possible that the early release of TFIIB breaks contacts to TFIIF, which is then improperly released.
TFIIB and TFIIF interact with numerous factors including the CTD phosphatases, Fcp1 (54) and Ssu72 (55, 56), that function after promoter escape when TFIIB has already been released. In addition, TFIIB is an essential component of chromosomal loops (56) that presumably must be reorganized after TFIIB is released during escape. One view is that the TFIIB tip region is at the center of an interaction network that breaks up with appropriate timing during escape, allowing proper linkage of transcription initiation to subsequent transcription processing events (18, 39). RNA polymerases that have used TFIIB in the context of this network (that is, at proper promoter sites) should retain the memory of correct initiation, meaning that RNA polymerases paused or elongating in the genome might be identifiable as having arisen from either correct or incorrect initiation events. This should be true whether the correct initiation events occurred during the pioneer round of transcription or subsequent rounds of facilitated reinitiation (31) via the promoter scaffold (9) and loop because all rounds require newly associated TFIIB. In this view the tip of TFIIB serves as a checkpoint monitor, directing only properly associated RNA polymerases to make functional mRNA.
Acknowledgments
We thank Michael Carey, Carol Eng, and Vivian Y. Shi for technical advice, Arnold J. Berk for the generous gift of HeLa cells, and Albert J. Courey, Yixin X. Huo, Harold G. Martinson, and Shakir Sayani for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM49048. This work was also supported by a grant from the UCLA Academic Senate.
- CTD
- C-terminal domain.
REFERENCES
- 1.Kornberg R. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12955–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratowski S., Zhou H. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5633–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran K., Gralla J. D. (2008) J. Biol. Chem. 283, 15665–15671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson N. E., Glaser B. T., Foley K. M., Burton Z. F., Burgess R. R. (2009) J. Biol. Chem. 284, 24754–24766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushnell D. A., Westover K. D., Davis R. E., Kornberg R. D. (2004) Science 303, 983–988 [DOI] [PubMed] [Google Scholar]
- 6.Liu X., Bushnell D. A., Wang D., Calero G., Kornberg R. D. (2010) Science 327, 206–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostrewa D., Zeller M. E., Armache K. J., Seizl M., Leike K., Thomm M., Cramer P. (2009) Nature 462, 323–330 [DOI] [PubMed] [Google Scholar]
- 8.Chi T., Lieberman P., Ellwood K., Carey M. (1995) Nature 377, 254–257 [DOI] [PubMed] [Google Scholar]
- 9.Yudkovsky N., Ranish J. A., Hahn S. (2000) Nature 408, 225–229 [DOI] [PubMed] [Google Scholar]
- 10.Dvir A. (2002) Biochim. Biophys. Acta 1577, 208–223 [DOI] [PubMed] [Google Scholar]
- 11.Buratowski S. (2009) Mol. Cell 36, 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandal S. S., Chu C., Wada T., Handa H., Shatkin A. J., Reinberg D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7572–7577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn S. H., Keogh M. C., Buratowski S. (2009) EMBO J. 28, 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Fairley J. A., Roberts S. G. (2010) Curr. Biol. 20, 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hieb A. R., Baran S., Goodrich J. A., Kugel J. F. (2006) EMBO J. 25, 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Almeida S. F., Carmo-Fonseca M. (2008) FEBS Lett. 582, 1971–1976 [DOI] [PubMed] [Google Scholar]
- 17.Trigon S., Serizawa H., Conaway J. W., Conaway R. C., Jackson S. P., Morange M. (1998) J. Biol. Chem. 273, 6769–6775 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez C. R., Cho E. J., Keogh M. C., Moore C. L., Greenleaf A. L., Buratowski S. (2000) Mol. Cell. Biol. 20, 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho C. K., Shuman S. (1999) Mol. Cell 3, 405–411 [DOI] [PubMed] [Google Scholar]
- 20.Pei Y., Hausmann S., Ho C. K., Schwer B., Shuman S. (2001) J. Biol. Chem. 276, 28075–28082 [DOI] [PubMed] [Google Scholar]
- 21.Schroeder S. C., Schwer B., Shuman S., Bentley D. (2000) Genes Dev. 14, 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Max T., Søgaard M., Svejstrup J. Q. (2007) J. Biol. Chem. 282, 14113–14120 [DOI] [PubMed] [Google Scholar]
- 23.Cho E. J., Kobor M. S., Kim M., Greenblatt J., Buratowski S. (2001) Genes Dev. 15, 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou M., Halanski M. A., Radonovich M. F., Kashanchi F., Peng J., Price D. H., Brady J. N. (2000) Mol. Cell. Biol. 20, 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eissenberg J. C., Shilatifard A., Dorokhov N., Michener D. E. (2007) Mol. Genet. Genomics 277, 101–114 [DOI] [PubMed] [Google Scholar]
- 26.Licatalosi D. D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J. B., Bentley D. L. (2002) Mol. Cell 9, 1101–1111 [DOI] [PubMed] [Google Scholar]
- 27.Chambers R. S., Wang B. Q., Burton Z. F., Dahmus M. E. (1995) J. Biol. Chem. 270, 14962–14969 [DOI] [PubMed] [Google Scholar]
- 28.Chen H. T., Hahn S. (2004) Cell 119, 169–180 [DOI] [PubMed] [Google Scholar]
- 29.Chafin D. R., Claussen T. J., Price D. H. (1991) J. Biol. Chem. 266, 9256–9262 [PubMed] [Google Scholar]
- 30.Zawel L., Kumar K. P., Reinberg D. (1995) Genes Dev. 9, 1479–1490 [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y., Gralla J. D. (1993) Mol. Cell. Biol. 13, 4572–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami K. S., Masuda S., Darst S. A. (2002) Science 296, 1280–1284 [DOI] [PubMed] [Google Scholar]
- 33.Kulbachinskiy A., Mustaev A. (2006) J. Biol. Chem. 281, 18273–18276 [DOI] [PubMed] [Google Scholar]
- 34.Choi W. S., Lin Y. C., Gralla J. D. (2004) J. Mol. Biol. 340, 981–989 [DOI] [PubMed] [Google Scholar]
- 35.Choi W. S., Yan M., Nusinow D., Gralla J. D. (2002) J. Mol. Biol. 319, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 36.Freire-Picos M. A., Krishnamurthy S., Sun Z. W., Hampsey M. (2005) Nucleic Acids Res. 33, 5045–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Q., Moreland R. J., Conaway J. W., Conaway R. C. (1999) J. Biol. Chem. 274, 35668–35675 [DOI] [PubMed] [Google Scholar]
- 38.Komarnitsky P., Cho E. J., Buratowski S. (2000) Genes Dev. 14, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho E. J., Takagi T., Moore C. R., Buratowski S. (1997) Genes Dev. 11, 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou M., Deng L., Kashanchi F., Brady J. N., Shatkin A. J., Kumar A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12666–12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y. C., Choi W. S., Gralla J. D. (2005) Nat. Struct. Mol. Biol. 12, 603–607 [DOI] [PubMed] [Google Scholar]
- 42.Vandenbroucke K., Robbens S., Vandepoele K., Inzé D., Van de Peer Y., Van Breusegem F. (2008) Mol. Biol. Evol. 25, 507–516 [DOI] [PubMed] [Google Scholar]
- 43.Chen D., Wilkinson C. R., Watt S., Penkett C. J., Toone W. M., Jones N., Bähler J. (2008) Mol. Biol. Cell 19, 308–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mutoh N., Kawabata M., Kitajima S. (2005) J. Biochem. 138, 797–804 [DOI] [PubMed] [Google Scholar]
- 45.Bentley D. L. (2005) Curr. Opin. Cell Biol. 17, 251–256 [DOI] [PubMed] [Google Scholar]
- 46.Pinto I., Ware D. E., Hampsey M. (1992) Cell 68, 977–988 [DOI] [PubMed] [Google Scholar]
- 47.Birney E., et al. (2007) Nature 447, 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandler D. W., Gralla J. (1981) Nucleic Acids Res. 9, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Székvölgyi L., Rákosy Z., Bálint B. L., Kókai E., Imre L., Vereb G., Bacsó Z., Goda K., Varga S., Balázs M., Dombrádi V., Nagy L., Szabó G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14964–14969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson A. L., Loeb L. A. (2001) Mutat. Res. 477, 7–21 [DOI] [PubMed] [Google Scholar]
- 51.Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 52.Eichner J., Chen H. T., Warfield L., Hahn S. (2010) EMBO J. 29, 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng B., Price D. H. (2007) J. Biol. Chem. 282, 21901–21912 [DOI] [PubMed] [Google Scholar]
- 54.Kobor M. S., Simon L. D., Omichinski J., Zhong G., Archambault J., Greenblatt J. (2000) Mol. Cell. Biol. 20, 7438–7449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun Z. W., Hampsey M. (1996) Mol. Cell. Biol. 16, 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh B. N., Hampsey M. (2007) Mol. Cell 27, 806–816 [DOI] [PubMed] [Google Scholar]