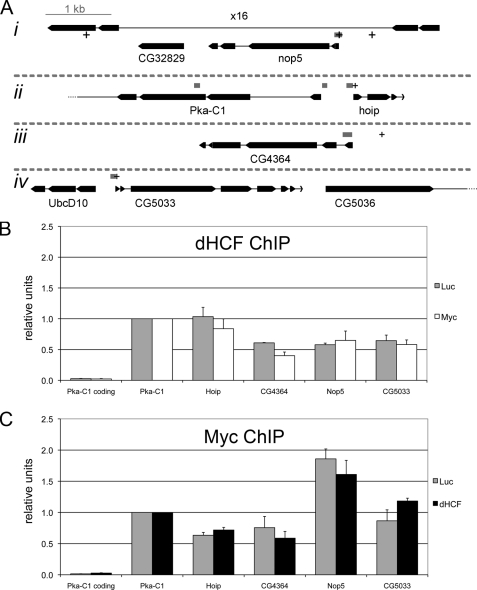
FIGURE 5.
dHCF and Myc co-localize at the promoters of Myc-regulated genes. A, schematic representation of the fragments probed in chromatin immunoprecipitations (ChIP assays). Exons are depicted as boxes and introns as thin lines. Gray boxes show positions of the PCR fragments and + signs consensus E-boxes. The fragment located in the coding region of Pka-C1 (3rd exon) serves as a negative control. All 4 chromosomal regions (i-iv) are depicted at the same scale. B and C, relative binding of dHCF (B) and Myc (C) to the indicated regions. SL2 cells were incubated with the indicated dsRNAs (Myc, dHCF, or Luciferase, which served as a negative control), and chromatin extracts prepared 3 days later for ChIP with polyclonal antibodies against dHCF or Myc. The diagrams represent the average of three biological replicates, and standard errors are shown as error bars.