Abstract
DNA gyrase is an essential bacterial enzyme required for the maintenance of chromosomal DNA topology. This enzyme is the target of several protein toxins encoded in toxin-antitoxin (TA) loci as well as of man-made antibiotics such as quinolones. The genome of Vibrio cholerae, the cause of cholera, contains three putative TA loci that exhibit modest similarity to the RK2 plasmid-borne parDE TA locus, which is thought to target gyrase although its mechanism of action is uncharacterized. Here we investigated the V. cholerae parDE2 locus. We found that this locus encodes a functional proteic TA pair that is active in Escherichia coli as well as V. cholerae. ParD2 co-purified with ParE2 and interacted with it directly. Unlike many other antitoxins, ParD2 could prevent but not reverse ParE2 toxicity. ParE2, like the unrelated F-encoded toxin CcdB and quinolones, targeted the GyrA subunit and stalled the DNA-gyrase cleavage complex. However, in contrast to other gyrase poisons, ParE2 toxicity required ATP, and it interfered with gyrase-dependent DNA supercoiling but not DNA relaxation. ParE2 did not bind GyrA fragments bound by CcdB and quinolones, and a set of strains resistant to a variety of known gyrase inhibitors all exhibited sensitivity to ParE2. Together, our findings suggest that ParE2 and presumably its many plasmid- and chromosome-encoded homologues inhibit gyrase in a different manner than previously described agents.
Keywords: Bacteria, Bacterial Toxins, DNA Gyrase, DNA Topoisomerase, DNA Topology
Introduction
Toxin-antitoxin (TA)3 loci encode a toxic protein (the toxin) and an antitoxin, either RNA or protein, that can neutralize the toxin activity. TA loci were originally identified in low copy plasmids (1, 2), where they ensure plasmid maintenance by killing plasmid-free daughter cells in a process known as post-segregational killing (3). Plasmid-borne TA loci promote plasmid maintenance because antitoxins are more labile than their cognate toxins in the intracellular environment. Plasmid-free cells, which can no longer synthesize either toxin or antitoxin, are therefore subject to the growth inhibitory/killing effect of the relatively long-lived toxin. Although TA loci were initially discovered in plasmids, in the current “post-genomic era,” it has become clear that these loci are also present in the chromosomes of nearly all species of bacteria, often in multiple copies (4). Some bacteria harbor very large numbers of TA loci in their chromosome (5), typically in association with mobile genetic elements. For example, in Vibrio cholerae, 13 TA loci are found associated with attC recombination sequences,which apparently enabled their capture by the integrase of the chromosome 2 superintegron (6). Even though the biochemical activities of several chromosome-encoded toxins have been deciphered, the physiologic significance of these ubiquitous loci remains the subject of controversy (7–9).
CcdB and ParE are representative of two families of unrelated toxins that block DNA replication by inhibiting DNA gyrase, an essential enzyme that is also the target of quinolone antibacterial agents (10). ParE, a toxin encoded on plasmid RK2 (11–13), and CcdB, a toxin encoded on the F plasmid, have unrelated amino acid sequences, but they both poison DNA gyrase. CcdB and ParE are encoded adjacent to proteic antitoxins, known as CcdA and ParD, respectively. Proteins similar to CcdB and ParE are encoded within plasmid and chromosomal sequences (4). Chromosomal CcdB homologues have been shown to target gyrase (14), but studies demonstrating that chromosomal ParE homologues poison this essential enzyme have not been reported.
Like other type II topoisomerases, DNA gyrase modifies DNA topology by introducing a double-stranded break in DNA through which a second DNA duplex is passed (15). This process can result in relaxation of positive or negative supercoils, both of which are energetically favored. Gyrase can also introduce negative supercoils; this process requires ATP. Both transcription and DNA replication generate positively supercoiled DNA, and gyrase is required to relieve the topological stresses associated with these essential processes. Maintenance of correct levels of chromosomal superhelicity is also critical for initiation of DNA replication and for the formation of open complexes for initiation of transcription (16).
Gyrase is a tetramer composed of two GyrA and two GyrB subunits, and both subunits contain distinct functional domains. The N-terminal domain of GyrA catalyzes the cleavage and rejoining of DNA, and its C-terminal domain binds and wraps DNA around the enzyme. Without the GyrA C-terminal domain (GyrA-CTD), also called the “DNA wrapping domain” or GyrA33 (17), gyrase is unable to negatively supercoil DNA; however, it still retains low levels of relaxation activity (18). The N-terminal domain of GyrB binds and hydrolyzes ATP, whereas its C-terminal domain interacts with GyrA and DNA (16, 19).
The mechanism of action of relatively few gyrase inhibitors has been determined. CcdB has been found to bind the dimerization domain of GyrA, thereby preventing strand passage as well as closure of the enzyme. In the presence of CcdB, the covalently linked DNA gyrase reaction intermediates are stabilized, which generates a “road block” for cellular polymerases in vivo and detectable DNA fragmentation in vitro (20). Quinolone antibiotics, such as nalidixic acid, also stabilize DNA gyrase intermediates, although they and CcdB target distinct sites within GyrA (16). ParE from plasmid RK2 is also thought to poison gyrase by stabilizing gyrase-DNA complexes, but the interactions between ParE and gyrase subunits and the mechanism by which ParE inhibits gyrase have not been explored. An Escherichia coli strain harboring a CcdB-resistant GyrA was not resistant to RK2-encoded ParE, raising the possibility that ParE inhibits gyrase in a different manner than CcdB (11). Putative ParDE homologues are encoded in the genomes of a wide variety of Gram-negative and Gram-positive bacteria (4, 21), but studies of the target and mechanisms of these chromosome-borne TA systems have not been conducted.
Here, we investigated the activities encoded by the parDE2 locus found in the V. cholerae superintegron. In this Gram-negative rod, the cause of cholera, the 13 putative TA loci include 3 loci with modest similarity to parDE of RK2. The predicted ParE2 amino acid sequence exhibits 29% sequence identity with RK2-ParE, whereas the predicted ParD2 sequence is only 12% identical to RK2 ParD. We found that the V. cholerae parDE2 genes encode a functional TA pair. Overexpression of ParE2 inhibited the growth of both V. cholerae and E. coli. ParD2 could prevent but not reverse ParE2 toxicity and, thus, may function in a different fashion than many other antitoxins. ParD2 co-purified along with ParE2, suggesting that ParD2-mediated neutralization of ParE2 toxicity results from the in vivo formation of a ParE2-ParD2 protein complex. Gyrase also co-purified with ParE2; in vitro studies revealed that distinct sites within GyrA are bound by ParE2 and CcdB. Unlike quinolones and CcdB, ParE2 requires ATP to stabilize gyrase-DNA cleavage complexes. In aggregate, our findings suggest that ParE2 inhibits gyrase in a different manner from other gyrase toxins.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
The plasmids and strains used in this study are listed in Table 1. All the gyrase mutant strains were generously provided by Coli Genetic Stock Center at Yale University. Bacteria were grown in LB medium at 37 °C. Antibiotics were used at the following concentrations: streptomycin, 200 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml for E. coli and 5 μg/ml for V. cholerae.
TABLE 1.
Strains and plasmids used in this study
| Name | Relevant description | Source or reference |
|---|---|---|
| Strains | ||
| N16961 | V. cholerae El Tor biotype | Ref. 26 |
| JY367 | N16961/pBAD33parE2+pGZparD2 | This study |
| JY370 | N16961/pBAD33parE2+pGZvector | This study |
| BW27784 | Ref. 28 | |
| JY 233 | BW27784/pBAD33parE2+pGZparD2 | This study |
| JY569 | BW27784/pBAD33parE2+pGZvector | This study |
| BL21(DE3) | Novagen | |
| JY262 | BL21(DE3)/pET28bHisparD2 | This study |
| JY302 | BL21(DE3)/pET28bHisparE2+pBAD33parD2 | This study |
| JY307 | BL21(DE3)/pET28bHisparD2+pBAD33parE2myc | This study |
| JY384 | BL21(DE3)/pET28bHisgyrA | This study |
| JY444 | BL21(DE3)/pET28bHisgyrB | This study |
| JY664 | BL21(DE3)/pET28bHisparD2+pBAD33ccdBv.f. | This study |
| JY668 | BL21(DE3)/pET28bHisgyrA14+pBAD33ccdBv.f. | This study |
| JY670 | BL21(DE3)/pET28bHisgyrA14+pBAD33parE2 | This study |
| JY281 | DH5α/pBAD33parE2myc | This study |
| Plasmids | ||
| pBAD33 | Ref. 53 | |
| BAD33parE2 | parE2 gene in SacI-XbaI sites of pBAD33 | This study |
| pBAD33ccdBv.f. | V. fischeri ccdB gene in SacI-XbaI sites of pBAD33 | This study |
| pGZ119EH | Ref. 54 | |
| pGZparD2 | parD2 gene in EcoRI-XbaI sites of pGZ119 | This study |
| pET28b | Ref. 22 | |
| pET28bparE2 | parE2 gene in NdeI-XhoI sites of pET28b | This study |
| pET28bparD2 | parD2 gene in NdeI-XhoI sites of pET28b | This study |
| pET28bgyrA | E. coli gyrA in NdeI-XhoI sites of pET28b | This study |
| pET28bgyrB | E. coli gyrB in NdeI-XhoI sites of pET28b | This study |
| pET28bgyrA14 | E. coli gyrA14 in NdeI-BamHI sites of pET28b | This study |
Mapping the 5′ End of the parD2 Transcript Analysis
Total RNA was extracted from N16961 using TRIzol (Invitrogen), and after DNase I (Qiagen) treatment, 5′ rapid amplification of cDNA ends was performed according to manufacturer's protocol (Invitrogen). The reverse transcription reaction was performed with JPP78 (TCATCGAATGTTTTCACTAT) and then PCR-amplified with the Abridged Anchor Primer (Invitrogen) and JPP80 (TGAAGCTATCAAGGTCATAATCAGCGTCA).
In Vivo ParE2 and ParD2 Activity Assays
V. cholerae and E. coli strains were grown in LB with appropriate antibiotics and 0.2% glucose to an A600 of ∼0.3–0.5, spun down, washed twice with LB and then resuspended in LB with antibiotics and either 0.2% glucose or 0.02% arabinose and grown at 37 °C. At successive time points, aliquots were removed and plated onto selective plates containing 0.2% glucose to enumerate colony-forming units (CFU).
Protein Purification
E. coli gyrA and gyrB and V. cholerae parD2 and parE2 were each introduced into a modified pET28b (+) expression vector (Novagen) to yield recombinant proteins that contain a His6 tag on their respective N termini. This modified pET28b vector encodes a tobacco etch virus protease recognition site (22). The DNA sequences of these constructs were confirmed. Proteins were expressed and purified according to a standard protocol (GE Healthcare). Briefly, 1-liter cultures of E. coli BL21(DE3) containing one of the expression vectors was grown at 37 °C in LB supplemented with kanamycin to an A600 of ∼0.6, then protein expression was induced by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside (IPTG), and growth was continued for an additional 4 h at 25 °C. Then the cells were lysed in a French pressure cell, and the His-tagged proteins were purified using His-Trap nickel columns (Amersham Biosciences) on an FPLC. The column was equilibrated with binding buffer (50 mm NaH2PO4, pH 8.0, 500 mm NaCl. 20 mm imidazole, 10% glycerol). After washing with buffer W (50 mm NaH2PO4, pH 8.0, 500 mm NaCl, 20 mm imidazole, 10% glycerol), His6-tagged proteins were eluted from the column with a 20-ml linear imidazole gradient (20–500 mm imidazole). Fractions containing pure protein (purity >90% estimated by SDS-PAGE) were pooled and dialyzed overnight at 4 °C in protein storage buffer (20 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 3 mm DTT, 50% glycerol). Size-exclusion chromatography was subsequently performed for further purification of His-GyrA and His-GyrB. When purifying His-tagged versions of either GyrB or GyrA, we noticed that endogenous GyrA or GyrB, respectively, always co-purified along with the epitope-tagged subunit. This was routinely monitored with dot blots performed with anti-GyrA and anti-GyrB antibodies. These blots revealed that more GyrA co-purified with His-GyrB than GyrB with His-GyrA (data not shown).
Native ParE2 was generated by removing the N-terminal His6 tag from purified His6-ParE2 using AcTEV protease (Invitrogen) according to manufacturer's protocol. Vibrio fischeri CcdB and E. coli GyrA14, GyrA59 were produced as described by Dao-Thi et al. (23) and De Jonge et al. (14), respectively. E. coli GyrB was provided by Inspiralis Ltd. (Norwich, UK).
In Vivo Protein Interaction and Analysis
Log-phase cultures of JY302 or JY307 were grown in 0.02% arabinose and 50 μm IPTG for 2 h to induce both His-ParE2 and ParD2-myc expression. His6ParE2 was affinity-purified using an Ni-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) according to the manufacturer's instructions. Briefly, cell lysates were incubated with Ni-NTA resin equilibrated with binding buffer 50 mm NaH2PO4, pH 8.0, 500 mm NaCl, 10 mm imidazole for 2 h at 4 °C. Wash buffer was 50 mm NaH2PO4, pH 8.0, 500 mm NaCl, 20 mm imidazole, and elution buffer was 50 mm NaH2PO4, pH 8.0, 500 mm NaCl, 250 mm imidazole. All fractions were collected and analyzed by Western blotting. Blots were probed with anti-Myc antibody (Invitrogen) or anti-His antibody (Genetex) or anti-GyrA antibody (Inspiralis) or anti-GyrB antibody (Inspiralis).
Surface Plasmon Resonance
Surface plasmon resonance experiments were performed on a BIAcore® 2000 system (GE Healthcare). The interactions between V. cholerae ParE2 or V. fischeri CcdB and various gyrase fragments were analyzed on both CM5 and Ni-NTA chips.
Sensor Chip CM5
E. coli His-GyrA, His-GyrA14, and His-GyrB were immobilized on a CM5 sensor chip (GE Healthcare) via amine coupling. For immobilization, the system was initially primed with Hepes-buffered saline with a flow rate of 5 μl/min. The carboxylated dextran matrix was activated by a 7-min injection of a solution containing 0.2 m N-ethyl-N′-(3-diethylamino)propyl) carbodiimide and 0.05 m N-hydroxysuccinimide. A protein solution in 10 mm sodium acetate, pH 4.5, was then injected until the desired amount of protein was immobilized. The surface immobilization was then blocked by a 7-min injection of 1 m ethanolamine hydrochloride. Hen-egg lysozyme was coupled in a similar fashion in the remaining flow cell for reference subtraction. After having been coupled to the CM5 chip, His-GyrB was washed with regenerating buffer (50 mm NaOH, 1 m NaOH) before analysis. This protocol greatly reduced the amount of the co-purifying untagged GyrA subunit (data not shown). Binding experiments were performed at 25 °C in running buffer (35 mm Tris-HCl, pH 7.5, 25 mm NaCl, 5 mm MgCl2, 5 mm dithiothreitol, 5% glycerol, 0.02% Tween 20) that was filtered and de-gassed before usage. The flow was set at 30 μl/min, and data were collected at 1 Hz. All analytes were dialyzed into the running buffer before analysis. Association was observed for 120 s, and dissociation was observed for 240 s. Regeneration was performed with 10 μl of regeneration buffer. The binding data were analyzed with the BIAevaluation 4.1 software (GE Healthcare), and the figures were generated with pro Fit 6.1.11 (Quantum Soft).
For the kinetic analysis on the ParE2-GyrA59 interaction (Fig. 5G), E. coli GyrA59 was immobilized on a CM5 sensor chip in a similar manner. About 1200 relative units of GyrA59 was immobilized in flow cell 2 of the chip and washed with regeneration buffer (100 mm NaOH). The surface in flow cell 1 was used as a reference and treated only with N-ethyl-N′-(3-diethylamino)propyl) carbodiimide, N-hydroxysuccinimide, and ethanolamine. Sensorgrams of different V. cholerae ParE2 concentrations (3.9 nm, 7.8 nm, 15.6 nm, 31.25 nm, 62.6 nm, 125 nm, 250 nm, 500 nm, 1 μm, 2 μm) plus a 0 concentration (injection of running buffer) were collected in duplicate. The flow was set at 5 μl/min, and data were collected at 2 Hz. This low flow rate was chosen based on the small quantity of ParE2 available; purifying large quantities of the protein was not feasible given its toxicity. Mass transport phenomena are likely at play under these conditions, and thus, the reported values for kon, koff, and KD should be regarded as apparent.
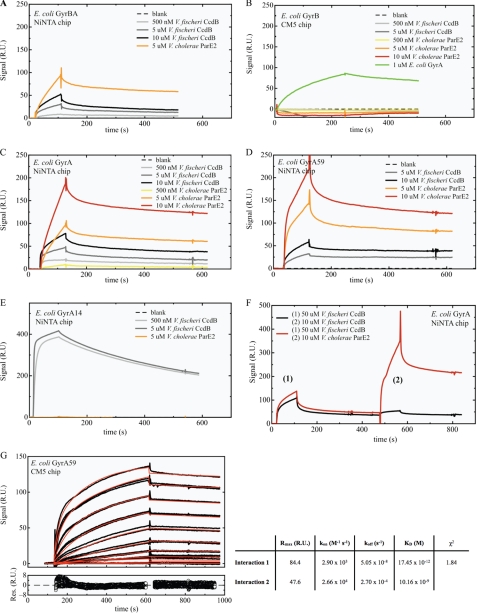
FIGURE 5.
SPR measurements of interactions between V. cholerae ParE2 or V. fischeri CcdB and gyrase. GyrBA (∼200 relative units (RU)) (A), GyrA (∼150 RU) (C), GyrA59 (∼600 RU) (D), and GyrA14 (∼400 RU) (E) were non-covalently coupled to a Ni-NTA chip, whereas GyrB (∼1800 RU) (B) was covalently coupled to a CM5 chip. The analytes were injected over the immobilized ligands at the indicated concentrations, expressed in terms of monomer. F, V. fischeri CcdB was injected at a concentration of 50 μm to saturate the CcdB binding site on GyrA. After a dissociation time of 200 s, ParE2 was injected at a concentration of 10 μm. A control experiment with a second CcdB (10 μm) injection showed that all CcdB binding sites are saturated after the first CcdB injection. G, shown is kinetic analysis of the interaction between V. cholerae ParE2 and E. coli GyrA59. The top graph displays the sensorgrams at different ParE2 concentrations (0 nm, 3.9 nm, 7.8 nm, 15.6 nm, 31.25 nm, 62.5 nm, 125 nm, 250 nm, 500 nm, 1 μm, 2 μm), which were collected in duplicate and are shown in black. The red lines represent the best fit of the model function (heterogeneous ligand model) to the experimental curves. The residuals of the fitting procedure are shown in the bottom graph. Model-based analyses of the sensorgrams recorded during the multicycle analysis of the ParE2-GyrA59 interaction indicates that the binding of ParE2 to GyrA59 is not monophasic and that two binding events occur in parallel. Fitting with a simple 1:1 Langmuir binding model does not result in acceptable residuals (χ2 = 22.4). The simplest model providing a reasonable fit to the data is a heterogeneous ligand model, resulting in a χ2 of 1.84. In aggregate, analysis of this data set reveals a very high affinity for the ParE2-GyrA59 interaction with a KD between ∼20 pm and 10 nm.
All analytes were dialyzed into the running buffer before analysis. Association was observed for 500 s and dissociation was observed for 300 s. Regeneration was performed with 10 μl of regeneration buffer. The sensorgrams were fitted to a heterogeneous ligand model according to the equations below after subtraction of the reference and zero concentration data, which provided the kinetic parameters for the ParE2-GyrA59 interaction.
In this model A denotes the analyte, and B denotes the ligand.
Sensor Chip NTA
E. coli GyrA, GyrA14, GyrA59, and GyrBA were immobilized on a BIAcore® Ni-NTA sensor chip (GE Healthcare) via their N-terminal hexahistidine tags as described (24). Binding experiments were performed as described above with a different running buffer (35 mm HEPES pH 7.5, 25 mm NaCl, 5 mm MgCl2, 0.005% Tween 20). To ensure that the chip was free of any ligand, 20 μl of regeneration buffer (10 mm HEPES, pH 8.3, 150 mm NaCl, 350 mm EDTA, 0.005% Tween 20) was passed over the flow cells at 5 μl/min. This was followed by a 20-μl injection of the activation buffer (500 μm NiSO4, 35 mm HEPES, pH 7.5, 25 mm NaCl, 5 mm MgCl2, 0.005% Tween 20) in flow cell 2 at 10 μl/min. The ligands His-GyrA14, His-GyrA59, His-GyrBA, and His-GyrA were, respectively, immobilized to the chip by injecting 10, 10, 10, and 50 μl of a 7 μg/ml solution at 20 μl/min followed by a dissociation time of 100 s. The analytes were injected at a volume of 30 μl at 20 μl/min followed by a dissociation time of at least 400 s. Regeneration was subsequently performed by injecting 40 μl of regeneration buffer at 10 μl/min.
For the interaction between V. cholerae ParD2 and ParE2, N-terminal His-tagged ParD2 was non-covalently coupled to a Ni-NTA chip by injecting 10 μl of a 7 μg/ml solution at 20 μl/min followed by a dissociation time of 100 s. The rest was performed as described for the ParE2-gyrase interaction on the Ni-NTA chip.
Gyrase Activity Assays
DNA gyrase supercoiling, relaxation, and cleavage assays were performed essentially as described (25). Substrates for these assays were 3.5 nm relaxed pBR322, 1.02 nm supercoiled pCB182, or 4.82 pm V. cholerae chromosomal DNA. For supercoiling and cleavage assays, the reaction buffer was 35 mm Tris-HCl, 24 mm KCl, 4 mm MgCl2, 2 mm DTT, 1.8 mm spermidine, 1 mm ATP, 6.5% glycerol, and 0.1 mg/ml BSA. For holoenzyme relaxation assays, ATP and spermidine were omitted from the assay buffer, and for A592B2 relaxation assays, ATP was included. 1 unit of DNA gyrase (30 nm) (New England Biolabs) or 100 nm A592B2, 1.88 μm ParE2, 2.25 μm ParD2, and 0.5 mm nalidixic acid (Sigma) were used as indicated. Both supercoiling and cleavage reactions were carried out at 37 °C for 1 h, whereas relaxation reactions were performed at 30 °C for 3.5 h. The reactions were stopped with 50 μg/ml proteinase K and 1% SDS. The DNA was precipitated with phenol:chloroform:isoamyl alcohol (25:24:1) (Invitrogen) before it was loaded on to 1% agarose gels and electrophoresed in the absence of ethidium bromide.
RESULTS
The V. cholerae parD2 Locus Was Mis-annotated
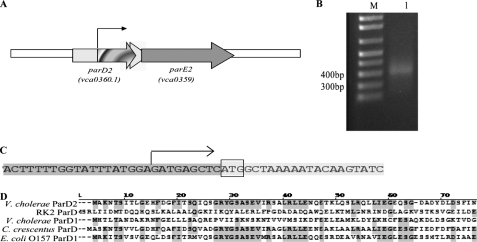
The predicted V. cholerae ParE2 amino acid sequence exhibits 29% sequence identity with RK2-ParE. In contrast, the predicted ParD2 sequence is only 12% identical to RK2 ParD. However, two different annotations of the V. cholerae parD2 locus have been reported. In the original annotation of the genome of V. cholerae El Tor strain N16961, the parD2 locus (vca0360) was predicted to encode a protein of 153 amino acids (26). In a bioinformatics study re-annotating microbial genomes, Bocs et al. (27) predicted that parD2 is actually a shorter ORF, vca0360.1, embedded within vca0360 (Fig. 1A). We used 5′ rapid amplification of cDNA ends to experimentally investigate the parD2 transcription start site. This analysis revealed that the start of parD2 transcription initiated 182 nucleotides 3′ of the annotated vca0360 translational start site, 11 bp 5′ of the translation start site predicted by Bocs et al. (27) (Fig. 1). Additional analyses suggest that there is a recognizable and functional promoter upstream of this transcription start site (data not shown). The predicted VCA0360.1 ORF yields a protein of 80 amino acids that exhibits up to 67% sequence similarity to other chromosome-encoded ParDs and 24% identity to the RK2 plasmid-encoded ParD (Fig. 1D). In contrast, the predicted VCA0360 amino acid sequence exhibited less similarity to other ParD2 proteins. Together, these observations strongly suggest that vca0360.1 encodes V. cholerae ParD2, and we used this parD2 annotation for the studies described below.
FIGURE 1.
Organization of the V. cholerae parDE2 locus. A, shown is a schematic representation of the parDE2 region in the V. cholerae N16961 chromosome. The white arrow represents the original vca0360 ORF as previously annotated (26), and the light gray arrow represents the vca0360.1 ORF as annotated in Bocs et al. (27). The thin arrow represents the predicted parD2 promoter based on the 5′ rapid amplification of cDNA ends. B: lane M, 1-kb plus marker; lane 1, specific PCR product from the 5′ rapid amplification of cDNA ends experiment. C, the start of the parD2 transcript, represented by the arrow, was determined by sequencing the PCR shown in B. The parD2 translational start site, as predicted by Bocs et al. (27), is shown with the box around the ATG. D, alignments of the predicted amino acid sequences of vca0360.1 (V. cholerae ParD2) with the indicated chromosome- and plasmid (RK2)-encoded ParD sequences are shown.
ParE2 Is Toxic to Both V. cholerae and E. coli, and Its Toxicity Can Be Ameliorated by ParD2
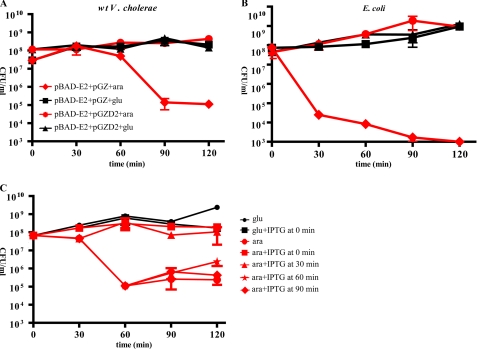
We constructed plasmid-borne inducible versions of parE2 and parD2 to test if these V. cholerae genes encode a functional toxin/antitoxin pair. Expression of parE2 from pBAD-E2 is induced by arabinose, whereas expression of parD2 from pGZ-D2 is constitutive and driven by its endogenous promoter. The parE2 expression construct was introduced along with either pGZ-D2 or a vector control into wild type El Tor V. cholerae strain N16961 and E. coli BW27784. Induction of parE2 expression in the absence of the parD2 expression construct was highly toxic in both strains. After parE2 induction, the number of CFU in cultures was reduced by ∼3–5 orders of magnitude relative to the number in cultures in which parE2 expression was repressed with glucose (Fig. 2, A and B, red diamonds versus black squares). The rapidity and magnitude of the reduction in CFU after induction of parE2 expression were even more dramatic in E. coli BW27784 than in V. cholerae, perhaps because this E. coli strain has a constitutive arabinose transporter and does not exhibit inducer exclusion (28), and ParD2 in the wild type V. cholerae strain may partially neutralize the effect of ParE2. Notably, no toxicity after parE2 induction was observed in any strain containing pGZ-D2. Collectively, these data suggest that parE2 encodes a toxin whose target is conserved between V. cholerae and E. coli and whose activity can be neutralized by ParD2.
FIGURE 2.
Effect of ParE2 and ParD2 on V. cholerae and E. coli viability. V. cholerae N16961 (A) and E. coli BW27784 (B) contained a plasmid-borne, arabinose-inducible parE2 (pBAD-E2) and either an empty vector (pGZ, diamonds and squares) or the vector harboring parD2 under control of its native promoter (pGZ-D2, circles and triangles). All cultures were grown in LB supplemented with 0.2% glucose at 37 °C until an A600 of ∼0.3, washed and resuspended in either LB plus 0.2% glucose (glu) (black squares and triangles) or LB plus 0.02% arabinose (ara) (red diamonds and circles). CFU were enumerated at the indicated time points. C, E. coli BL21 (pBAD-E2, pET-D2), (JY307), harbors plasmids with arabinose-inducible parE2 and IPTG-inducible parD2. Cultures were grown in LB supplemented with 0.2% glucose until an A600 of ∼0.3, washed and resuspended in either LB plus 0.2% glucose (glu) or glucose plus 50 μm IPTG (black squares and circles) or LB plus 0.02% arabinose (ara) (red shapes). IPTG (50 μm) was added at the indicated time points after the addition of arabinose. CFU were enumerated at the indicated time points.
We also tested whether ParD2 could reverse ParE2 toxicity in vivo. For these experiments we used E. coli BL21 containing pBAD-E2 and pET-parD2 (JY307), which enabled inducible expression of both proteins. The strain was initially cultured in glucose to repress parE2 expression. As described above, induction of parE2 expression in the absence of parD2 induction reduced the plating efficiency of this strain by several orders of magnitude, whereas induction of parE2 expression when parD2 was produced simultaneously did not impair plating efficiency (Fig. 2C). Induction of parD2 expression 30 min after induction of parE2 also did not reduce the number of CFU. However, when parD2 expression was initiated 60 or 90 min after induction of parE2, little or no neutralization of ParE2 toxicity was detected (Fig. 2C, red stars and red diamonds, respectively). These results suggest that ParD2, unlike several previously characterized antitoxins, such as RelE (29), may be unable to reverse the toxicity of its cognate toxin.
ParE2 and ParD2 Interact
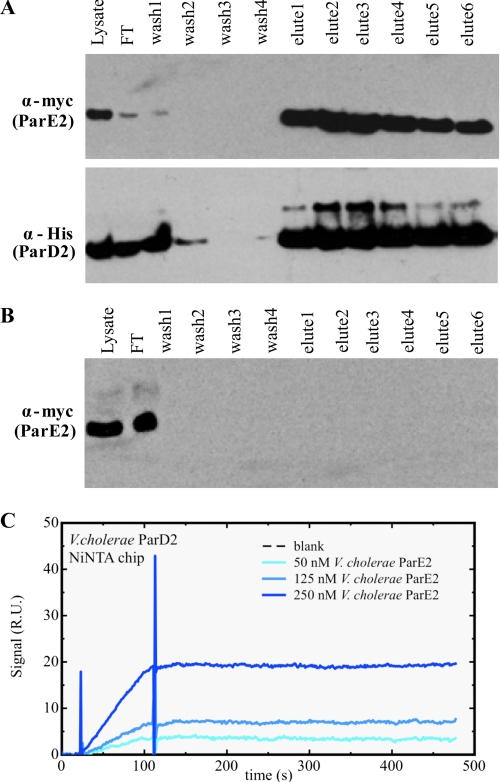
Protein antitoxins typically neutralize their cognate toxins by binding to them and inhibiting their ability to interact with their respective cellular targets (30–33), We investigated whether ParD2 interacts with ParE2 in vivo by testing whether epitope-tagged versions of these proteins would co-purify from E. coli BL21 co-expressing ParE2-Myc and His-ParD2 (JY307). The N-terminal His-tagged ParD2 was affinity-purified from lysates of JY307 using Ni-NTA resin. After washing, protein bound to the resin was eluted with increasing concentrations of imidazole. The eluted fractions were then immunoblotted with anti-His and anti-Myc antisera. As seen in Fig. 3A, these Western blots revealed that ParE2-Myc was purified along with His-ParD2. A mock purification using lysate from cells expressing ParE2-Myc but not His-ParD2 revealed that ParE2-Myc did not bind to the Ni-NTA resin in the absence of His-ParD2 (Fig. 3B). Together, these observations suggest that ParE2 is present in a complex containing ParD2, but they do not establish that there is direct contact between these proteins. However, surface plasmon resonance (SPR) assays using purified His-ParD2 and untagged ParE2 demonstrated that these proteins physically interact and form a stable complex (Fig. 3C).
FIGURE 3.
Co-purification of Myc tagged ParE2 with His-tagged ParD2. A, His-ParD2 from JY307, E. coli BL21 co-expressing His-ParD2 and ParE2-Myc (lysate) was affinity-purified using Ni-NTA resin. The starting lysate flowthrough (FT), washes, and eluted fractions were analyzed by Western blotting with anti-His and anti-Myc antibodies. B, cell lysate from JY281, E. coli DH5α-expressing ParE2-Myc were processed as in A and then analyzed using anti-Myc antibody. C, His-tagged ParD2 (∼50 relative units (R.U.) was coupled to a Ni-NTA chip, and native ParE2 was injected at the indicated concentrations, expressed in terms of monomer.
ParE2 Interacts with DNA Gyrase
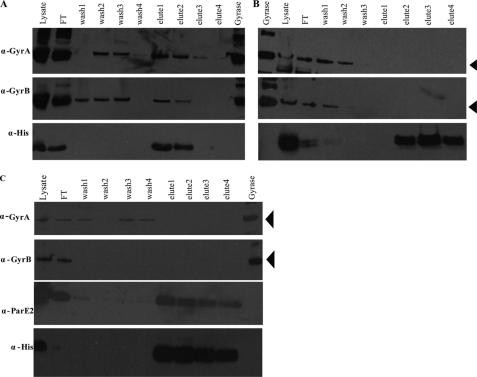
Even though ParE2 only exhibits a modest degree of sequence identity to the ParE encoded by plasmid RK2, we explored whether it has the same cellular target as the plasmid-encoded toxin DNA gyrase. First, we tested whether we could detect an association between ParE2 and gyrase in vivo, as described above for His-ParD2 and ParE2-Myc. Western blotting of proteins purified from E. coli BL21 expressing His-ParE2 (JY302) revealed that fractions containing affinity-purified His-ParE2 also contained GyrA and GyrB (Fig. 4A). GyrA and GyrB were not detected in control purifications performed using lysates of BL21-expressing His-ParD2 (JY262), indicating that they do not bind non-specifically to the affinity matrix (Fig. 4B). These results suggest that ParE2 is part of a complex containing GyrA and GyrB in vivo. Because GyrA and GyrB form a tight tetramer, it is not surprising that both gyrase subunits co-eluted with ParE2.
FIGURE 4.
Co-purification of GyrA and GyrB with His-ParE2. Cell lysates from E. coli BL21(DE3) expressing His-ParE2 (JY302) (A) or expressing His-ParD2(JY262) (B) or E. coli BL21 co-expressing His-ParD2 and ParE2-Myc (JY307) (C) were incubated with Ni-NTA resin, washed, and then eluted with imidazole. The initial lysates flowthrough (FT), washes, and eluted fractions were electrophoresed on polyacrylamide gels and then analyzed by Western blotting with anti-GyrA, anti-GyrB anti-His, and anti-ParE2 antibodies as indicated. Purified E. coli gyrase was used as the control. The positions of GyrA or GyrB are indicated with arrows.
To explore if ParD2 could prevent ParE2 interactions with gyrase, we overexpressed His-ParD2 along with ParE2. In this context, no GyrA or GyrB co-purified along with ParD2 and ParE2 (Fig. 4C), suggesting that when ParE2 is bound to ParD2, the toxin cannot interact with gyrase.
ParE2 Binds to GyrA
To further characterize the interactions of ParE2 and gyrase, the binding of ParE2 to gyrase and the individual gyrase subunits was probed using SPR. In these experiments we compared the interaction of ParE2 with different regions of DNA gyrase from E. coli immobilized on BIAcore sensor chips. As a control, we examined the binding of the V. fischeri CcdB toxin, which is known to bind to GyrA (34, 35), to the same chips. Initially, the binding of the toxins to GyrBA was examined. A His-GyrB-GyrA complex was non-covalently coupled to a Ni-NTA sensor chip to probe the interactions between the toxins and GyrBA. As seen in Fig. 5A, V. fischeri CcdB and V. cholerae ParE2 both bound to GyrBA.
Next, interactions between ParE2 and the individual gyrase subunits were examined. As shown in Fig. 5B, neither ParE2 nor CcdB interacted with GyrB alone. However, an association between GyrB and GyrA was observed (Fig. 5B, green line), suggesting that the GyrB bound to the chip was properly folded. In addition, we observed that ParE2, like CcdB, bound to chips coated only with His-GyrA (Fig. 5C). Together, these data indicate that the copurification of GyrB along with affinity-purified ParE2 (Fig. 4) was due to an indirect association of these proteins that was mediated by GyrA.
ParE2 Binds to a Different Site on GyrA than CcdB
To gain insight into the ParE2 target within GyrA, we made use of previously characterized fragments of this enzyme. We observed that ParE2, like CcdB, binds to GyrA59, a 59-kDa fragment that includes the N-terminal amino acid residues 2–523 but lacks the C-terminal DNA-wrapping domain (Fig. 5D). Model-based analysis of the sensorgrams recorded during the multicycle analysis of the ParE2-GyrA59 interaction revealed that the binding of ParE2 to GyrA59 is not monophasic; instead, two binding events occur in parallel. The simplest model providing a reasonable fit to the data is a heterogeneous ligand model. This model has a χ2 of 1.84 and indicates a high affinity for the ParE2-GyrA59 interaction with a KD between ∼20 pm and 10 nm (Fig. 5G). The heterogeneity in binding of ParE2 by GyrA59 was most likely introduced by the immobilization method. Such heterogeneity was also observed in the interaction of covalently coupled GyrA59 with CcdB (51).
Although ParE2 and CcdB both bind to GyrA59 with similar high affinity, the toxins differed in their ability to bind a GyrA fragment known as GyrA14 (amino acids 363–497), which mediates the dimerization of GyrA monomers. SPR revealed no interaction between ParE2 and this fragment, whereas, as previously reported, binding of CcdB was apparent, indicating that the two toxins recognize distinct regions within GyrA (31, 36, 37) (Fig. 5E). Further support for this conclusion was provided by the observation that a Ni-NTA-GyrA chip whose binding to CcdB is fully saturated can still readily bind ParE2 (Fig. 5F).
Furthermore, in vivo studies also indicated that ParE2 and CcdB have distinct targets within GyrA. Consistent with previous reports, expression of GyrA14 protected cells from the toxicity of CcdB, likely by titrating CcdB from its cellular target (37) (supplemental Fig. S1). In contrast, GyrA14 did not prevent ParE2 toxicity. Even in the presence of GyrA14, more than a 10-fold reduction in CFU was observed after induction of ParE2 expression (supplemental Fig. S1A). These findings provide strong support for the idea that ParE2 binds to a different domain of GyrA than CcdB. As expected, ParE2 toxicity was neutralized by ParD2 in this system, but ParD2 did not protect cells from the toxicity of V. fischeri CcdB (supplemental Fig. S1B).
ParE2 Stabilizes Gyrase-DNA Cleavage Intermediates
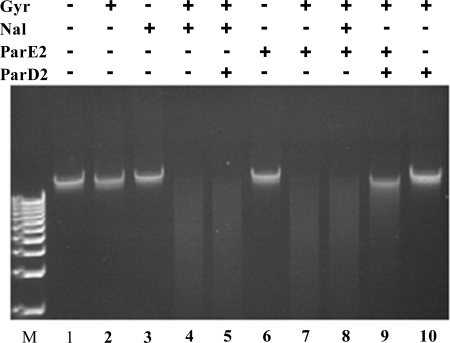
Studies on quinolone antibiotics (e.g. nalidixic acid (Nal)), CcdB, and RK2-encoded ParE revealed that these agents all inhibit the gyrase catalytic cycle by stabilizing DNA-gyrase cleavage intermediates. Such cleaved intermediates are readily detectable on agarose gels after reaction mixtures of gyrase, DNA, and toxins are treated with SDS and proteinase K (38, 39). We used a similar protocol to test whether ParE2 also stalls the gyrase-DNA cleavage complex using purified E. coli gyrase, ParE2, and V. cholerae chromosomal DNA as a substrate. In this assay Nal caused gyrase-dependent cleavage of the chromosomal DNA substrate (Fig. 6, lanes 3 and 4), as expected. The addition of ParE2 to gyrase also resulted in the formation of cleaved DNA (Fig. 6, compare lane 2 versus 7), but ParE2 did not cleave the DNA on its own (Fig. 6, lane 6). Together, these observations strongly suggest that ParE2, like Nal, stabilizes gyrase-DNA cleavage complexes. Similar results were also obtained when negatively supercoiled plasmid DNA was used as the substrate (data not shown).
FIGURE 6.
Effect of ParE2 and ParD2 on cleavage of chromosomal DNA by gyrase. In vitro reactions with chromosomal DNA, gyrase, ParE2, and ParD2 were carried out as described under “Experimental Procedures.” M indicates 1 kb plus DNA ladder.
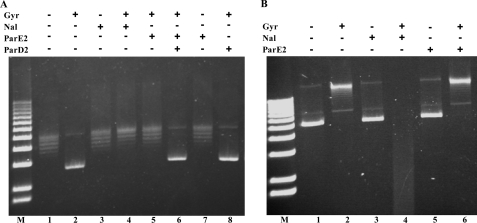
We carried out similar assays to test whether ParD2 could prevent and/or reverse ParE2 stabilization of gyrase-DNA cleavage complexes. When ParD2 was mixed with ParE2 before their addition to gyrase and DNA, near complete neutralization of ParE2 activity was observed (Fig. 6, compare lane 9 versus 7, and supplemental Fig. S2, lane 8). However, when ParD2 was added simultaneously with ParE2 to the reaction, less complete neutralization was observed (supplemental Fig. S2, lane 6). Finally, when ParD2 was added 30 min after ParE2, relatively little neutralization of the ParE2 effect was detected (supplemental Fig. S2, lane 7). We also found that ParE2-GyrA59 complex immobilized on the sensor chip could not be disrupted by ParD2 (data not shown). Together, these findings support the idea that ParD2 is unable to rescue ParE2-poisoned gyrase, which are consistent with the observation that ParD2 could not reverse ParE2 toxicity in vivo (Fig. 2C). Thus, ParD2 appears to differ from RK2 ParD (13) and CcdA (25), which can rejuvenate ParE- or CcdB-poisoned gyrase, respectively.
ParE2 Inhibits Gyrase Supercoiling Activity in an ATP-dependent Manner
We further characterized the ParE2 effect on gyrase utilizing additional in vitro assays of gyrase activity, as earlier studies of RK2 ParE did not specifically address whether this plasmid-encoded toxin inhibited gyrase-mediated DNA relaxation and/or supercoiling (13). Gyrase has previously been shown to relax supercoiled plasmid DNA and to introduce supercoils into relaxed plasmids in vitro. The first reaction proceeds relatively slowly but is ATP-independent, whereas the latter reaction requires ATP (25, 38, 39). Both can be interrupted by agents such as CcdB and quinolone antibiotics, which stabilize gyrase-DNA reaction intermediates.
We used plasmid DNA substrates and purified E. coli gyrase to investigate whether ParE2 interferes with gyrase-catalyzed supercoiling and/or relaxation of DNA. Using a relaxed plasmid substrate, we found that ParE2, like Nal and CcdB, inhibited introduction of supercoils by gyrase with an IC50 of ∼1.2 μm (Fig. 7A, compare lanes 2, 4, and 5, and supplemental Fig. S3), a similar value as CcdB (25). ParE2 effect could be blocked by preincubation of this protein with ParD2 (Fig. 7A, lane 8), consistent with the ParD2 previously noted role as an antitoxin in vivo and in vitro. In contrast, ParE2 and Nal had distinct effects upon relaxation of plasmid DNA (Fig. 7B and supplemental Fig. S4). The addition of a range of ParE2 concentrations had no effect upon gyrase-mediated relaxation of a supercoiled substrate (Fig. 7B, compare lane 2 and 6, and supplemental Fig. S4, compare lanes 3–7), whereas the addition of Nal (Fig. 7B, lane 4) resulted in accumulation of cleaved DNA intermediates (Fig. 7B, lane 4), as previously described (40). Furthermore, no or minimal inhibition of DNA relaxation was detected during a time course extending from 0 to 6 h in the presence of ParE2 (data not shown). These data suggest that ParE2, like Nal, can inhibit gyrase activity but that its mechanism of action differs. In particular, ParE2 appears to lack the capacity to block gyrase-mediated relaxation of DNA.
FIGURE 7.
The effect of ParE2 on gyrase supercoiling (A) and relaxation (B) of plasmid DNA substrates. The reactions were performed as described under “Experimental Procedures” except in B the reaction was stopped with SDS and proteinase K. In A, the substrate was relaxed pBR322 DNA, and in B the substrate was negatively supercoiled pCB182. M indicates 1Kb plus DNA ladder.
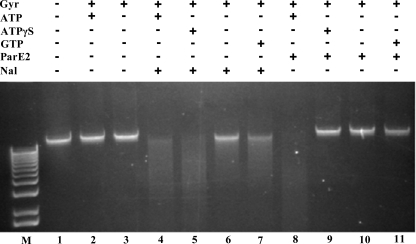
As the supercoiling assay, unlike the relaxation assay, is performed in the presence of ATP, we assessed whether the presence of nucleotide was important for ParE2 activity. For these assays we used a chromosomal DNA substrate (which is expected to mostly consist of linear DNA molecules). The addition of gyrase alone to this substrate either in the absence or presence of ATP had no discernible effect on its integrity, as expected when gyrase-mediated cleavage is followed by religation (Fig. 8). The addition of Nal resulted in accumulation of cleavage intermediates; as previously reported (38), this disruption of the gyrase reaction cycle was not dependent on ATP hydrolysis, although it did appear to be enhanced by the presence of nucleotide (Fig. 8, compare lanes 4–7). In contrast, the addition of ParE2 to reactions containing gyrase resulted in accumulation of cleavage intermediates only in the presence of ATP. No cleavage was detected when ATP was replaced by the non-hydrolyzable analog ATPγS or by GTP. Thus, although ParE2, like Nal and other gyrase poisons, can stabilize DNA-gyrase cleavage intermediates, the ParE2 effect appeared to be dependent upon gyrase binding to and hydrolyzing ATP.
FIGURE 8.
Requirement for ATP hydrolysis in ParE2 stabilization of gyrase-DNA cleavages. The reactions were carried out as described under “Experimental Procedures” using V. cholerae chromosomal DNA as substrate. 1 mm ATP, ATPγS, or GTP were used in these reactions. 5 μm ParE2 or 1 mm nalidixic acid were included as indicated. M indicates 1 kb plus DNA ladder.
An alternative explanation of the requirement for ATP for ParE2 toxicity is that the activity of ParE2 itself could depend on ATP. To exclude this possibility, we used a reconstituted gyrase (A592B2) containing a truncated GyrA that lacks the C-terminal 33-kDa DNA-wrapping domain of GyrA (18). The A592B2 complex cannot supercoil DNA, even in the presence of ATP, but it retains the ability to relax negatively supercoiled DNA. Consequently, use of this enzyme allowed the effect of ParE2 on relaxation of a supercoiled substrate to be assessed in the presence of ATP (17). In this assay, the A592B2 complex could convert nearly all of a supercoiled plasmid to a relaxed form (supplemental Fig. S5, A, lane 1 versus 2, and B, lane 1 versus 3). Inclusion of Nal in the reaction markedly inhibited A592B2 relaxation of the plasmid (supplemental Fig. S5A, lanes 3 and 4), similar to previous reports where ciprofloxacin was used (25). In contrast, a range of ParE2 concentrations did not inhibit A592B2 activity even in the presence of ATP (supplemental Fig. S5 A, lane 6, and B, lanes 4–7). These findings buttress the conclusion that ParE2 lacks the ability to block gyrase-mediated relaxation of DNA supercoils and argue against the possibility that ATP is required to convert ParE2 to an active form. Furthermore, these findings provide additional evidence that the ParE2 mechanism is distinct from those of quinolones and CcdB, both of which inhibit A592B2 activity (25).
Gyrase Mutants That Confer Resistance to Known Gyrase Toxins Do Not Confer Resistance to ParE2
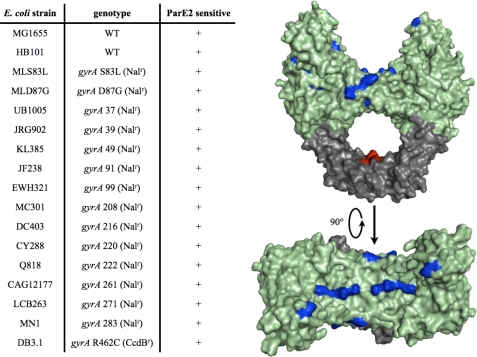
To continue to explore how ParE2 inhibits gyrase, we expressed ParE2 in a variety of E. coli strains harboring gyrase alleles that confer resistance to antibiotics and toxins that poison this enzyme. Strains harboring gyrB mutations conferring resistance to coumermycin A1 and microcin B17 were sensitive to ParE2 (data not shown). This was an expected result, as our findings indicated that ParE2 targets GyrA. Our observations presented above also suggest that ParE2 targets a different part of GyrA than CcdB. Consistent with this idea, DB3.1, a strain containing the CcdB-resistant gyrA R462C, was also sensitive to ParE2 (Fig. 9). Finally, all of the strains harboring gyrA alleles conferring resistance to Nal were sensitive to ParE2. In aggregate, these observations suggest that ParE2 poisons gyrase via a mechanism that is distinct from previously described agents that target this essential enzyme.
FIGURE 9.
Strains bearing gyrase alleles conferring resistance to several gyrase toxins remain sensitive to ParE2. In A, the strains indicated were tested for their resistance to ParE2 after the toxin was expressed from pBAD-E2. Strains were scored as sensitive if there was attenuated growth after induction of ParE2 expression with arabinose. In B, the mutations listed in A were mapped onto the crystal structure of E. coli GyrA59 colored in green (PDB entry 1AB4 (52). GyrA14 is shown in dark gray, and the red patches correspond to the residues involved in the interaction between CcdB and GyrA (Gln-456, Asp-460, Arg-462, Gln-464). The residues conferring resistance to quinolones when mutated are colored blue. This figure was generated using MacPyMOL (DeLano Scientific, Ltd.).
DISCUSSION
Our findings revealed that the parDE2 locus found in the superintegron of V. cholerae chromosome II encodes a functional proteic TA pair. Overexpression of ParE2 markedly inhibited growth of V. cholerae as well as E. coli, indicating that ParE2 toxicity is not restricted to V. cholerae and that the cellular target of ParE2 is conserved in both organisms. The parD2 ORF was mis-annotated in the V. cholerae genome, but expression of the correct parD2 ORF neutralized ParE2 toxicity. ParD2 co-purified with ParE2, and direct interactions between the proteins were detected with SPR. However, ParD2 appears to function in a different fashion than many other antitoxins, as ParD2 could prevent but not reverse ParE2 toxicity. ParE2, like plasmid RK2-encoded ParE, CcdB, microcin B17, and quinolone and coumarin antibiotics, targets DNA gyrase. We found that ParE2, like CcdB and quinolones, targets GyrA and stabilizes the DNA-gyrase cleavage complex. However, ParE2, in contrast to quinolones and CcdB, did not bind to GyrA14 and required ATP hydrolysis to stabilize gyrase cleavage intermediates. Also unlike quinolones and CcdB, ParE2 only interferes with gyrase supercoiling; its ability to relax DNA is apparently unaffected. Together, these findings suggest that ParE2 may only gain access to its target site(s) on GyrA at a particular stage of the supercoiling reaction, possibly after DNA wrapping and ATP hydrolysis. Finally, a set of strains resistant to a variety of known gyrase inhibitors all exhibited sensitivity to ParE2. Thus, taken together, our findings strongly suggest that ParE2 and most likely its many plasmid- and chromosome-encoded homologues inhibit gyrase in a different manner than previously described agents that poison this essential enzyme.
ParD2 Action and Its Physiological Implications
The interaction of ParD2 with ParE2 appears to be unusual. Our findings indicate ParD2, like other antitoxins, binds to its cognate toxin ParE2. However, unlike several antitoxins, including RK2 ParD, and F-plasmid CcdA (13, 41), V. cholerae ParD2 appears to be unable to reverse ParE2 inhibition of gyrase activity (Fig. 6). Structural studies have revealed that at least some antitoxins interact with their cognate toxins through direct binding of a disordered region present in the antitoxin C termini. As a consequence of such interactions, the disordered region reorganizes into a well defined structure, and conformational changes occur in the toxin, distorting the binding site for the cellular target (42–44). Recently, the crystal structure of the Caulobacter crescentus ParD1-ParE1 complex was reported (45). Based on this structure, Dalton and Crosson (45) proposed that ParD interactions with ParE do not induce large conformational changes in the toxin. Because C. crescentus ParD and V. cholerae ParD2 are similar (50% identity), we suspect ParD2 and ParE2 interact in a similar manner as their C. crescentus homologues. If this is the case, it is tempting to propose that the inability of ParD2 to reverse ParE2 toxicity stems from the absence of significant changes in ParE2 structure when it is bound by ParD2. Thus, if ParE2 is bound to gyrase, ParD2 binding may not lead to release gyrase from ParE2. A challenge for future studies will be to define if and how ParD2 prevents ParE2 interaction with GyrA in vivo. Recently Hallez et al. (46) reported that PaaA, an antitoxin for a ParE homologue in E. coli O157, altered the toxin subcelluar localization. It is possible that ParD2, which lacks similarity to PaaA, blocks ParE2 toxicity in a similar fashion as PaaA by sequestering the toxin away from gyrase.
Although there is considerable controversy regarding the physiologic function of chromosomal TA loci, previous work has led to the suggestion that such loci contribute to genome integrity. For example, two Vibrio vulnificus superintegron-encoded TA loci (relBE1 and parDE1) appear to prevent large scale chromosome loss (6). The parDE2 locus and likely the two parDE1 loci in the V. cholerae chromosome II superintegron may also function to promote the integrity of the V. cholerae genome, which like all members of the Vibrionaceae is divided between two chromosomes. V. cholerae strains that lose chromosome II undergo a characteristic series of cytologic changes culminating in cell death (47). One of the phenotypes observed after chromosome II loss is the guillotining of the remaining chromosome I. The DNA damage observed in the cells lacking chromosome II resembles the catastrophic effect of ParE2 on V. cholerae chromosomal DNA shown in Fig. 6. Thus, parDE2 might contribute to V. cholerae genome integrity via a mechanism similar to post-segregational killing. After loss of chromosome II (and the parDE2 locus), the more stable ParE2 would outlive the ParD2 antitoxin and poison DNA gyrase leading to the cleavage of chromosome I. We are currently exploring whether the V. cholerae parDE loci contribute to destruction of chromosome I when chromosome II is lost from cells.
A Novel Mechanism for Poisoning Gyrase
Our findings suggest that ParE2 interacts with gyrase at a site different from that of other agents that target this enzyme (Fig. 9). Even though we found that, like CcdB (51, 55), ParE2 binds to GyrA59 with high affinity (Fig. 5), ParE2 did not interact with GyrA14 (Fig. 5), unlike the CcdB family of toxins. Furthermore, ParE2 bound to GyrA saturated with CcdB (Fig. 5F). The locations of the residues conferring CcdB and Nal resistance are mapped on to the crystal structure of GyrA59 in Fig. 9. Because these residues are mostly found in the core of the GyrA subunit, it is likely that ParE2 interacts somewhere else on this gyrase subunit.
We found that ParE2, like CcdB and quinolones, can stabilize gyrase-DNA cleavage intermediates (Fig. 6). Covalent gyrase-DNA intermediates are formed during the gyrase catalytic cycle after the enzyme binds and cleaves the “G segment” of the DNA duplex (48). Ordinarily (in the absence of toxin), gyrase then transports the “T segment” through the resulting “gate” and then reseals the gap, thereby altering DNA topology. In contrast to quinolones and CcdB, ATP is required for ParE2 to stall the gyrase cleavage complex (Fig. 8). Although the precise role of ATP in the gyrase cycle is still not clear, it is generally accepted that its binding and hydrolysis trigger conformational changes in the entire enzyme complex (19). Upon binding ATP, the N-terminal ATPase domains of GyrB dimerize and close the “ATP-operated clamp,” capturing the T segment, which is then passed through the transiently cleaved G-segment. It has been suggested that ATP hydrolysis provides the driving force for transport of the T segment, when it passes from the “top” to the “bottom” of the enzyme (49). The latter step is thought to be essential for the gyrase supercoiling cycle (19). Thus, it is possible that changes in gyrase structure that accompany DNA wrapping and ATP hydrolysis, such as T segment top-down passage and the opening of the DNA gate, are required for ParE2 to interact with gyrase and inhibit its supercoiling activity. Because similar conformational changes in gyrase structure likely do not occur during enzyme relaxation of DNA, which proceed in the absence of ATP and DNA wrapping (19, 50), ParE2 is incapable of inhibiting gyrase relaxation activity. We nevertheless detected ParE2 binding to gyrase on the chip in the absence of ATP. However, previous studies have shown that gyrase can adopt atypical conformations when bound to SPR chips (51). Additionally, it is possible that ParE2 gyrase interactions are ordinarily constrained by the presence of DNA in supercoiling and relaxation assays.
Overall, the action of ParE2 on gyrase resembles the action of CcdB. However, there clearly are differences; for example, the requirement of ATP for ParE2 toxic action. It is likely that the two toxins do not share the same binding site, as the GyrA14 peptide did not abrogate the toxic action of ParE2. Given the extensive nature of the GyrA-GyrA interface that is likely to be transiently revealed during the ATP-dependent supercoiling process, we suggest that ParE2 may bind somewhere in this interface region. This is a topic for future investigation.
Supplementary Material
Acknowledgments
We thank Dr. Brigid Davis and Monica P. Hui for helpful discussions and comments on manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R37 AI-42347. This work was also supported by the Howard Hughes Medical Institute (to M. K. W.). Part of this work was supported by grants from Vlaams Instituut voor Biotechnologie, Vrije Universiteit Brussel and FWO-Vlaanderen. This work was also supported by the Biotechnology and Biological Sciences Research Council, Swindon, United Kingdom (to A. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- TA
- toxin-antitoxin
- CFU
- colony-forming units
- IPTG
- isopropyl-β-d-thiogalactopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- SPR
- surface plasmon resonance
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- Nal
- nalidixic acid.
REFERENCES
- 1.Jensen R. B., Gerdes K. (1995) Mol. Microbiol. 17, 205–210 [DOI] [PubMed] [Google Scholar]
- 2.Ogura T., Hiraga S. (1983) Cell 32, 351–360 [DOI] [PubMed] [Google Scholar]
- 3.Buts L., Lah J., Dao-Thi M. H., Wyns L., Loris R. (2005) Trends Biochem. Sci. 30, 672–679 [DOI] [PubMed] [Google Scholar]
- 4.Pandey D. P., Gerdes K. (2005) Nucleic Acids Res. 33, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramage H. R., Connolly L. E., Cox J. S. (2009) PLoS Genet 5, e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szekeres S., Dauti M., Wilde C., Mazel D., Rowe-Magnus D. A. (2007) Mol. Microbiol. 63, 1588–1605 [DOI] [PubMed] [Google Scholar]
- 7.Engelberg-Kulka H., Amitai S., Kolodkin-Gal I., Hazan R. (2006) PLoS Genet. 2, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsilibaris V., Maenhaut-Michel G., Mine N., Van Melderen L. (2007) J. Bacteriol. 189, 6101–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Melderen L., Saavedra De Bast M. (2009) PLoS Genet 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maxwell A. (1997) Trends Microbiol. 5, 102–109 [DOI] [PubMed] [Google Scholar]
- 11.Roberts R. C., Ström A. R., Helinski D. R. (1994) J. Mol. Biol. 237, 35–51 [DOI] [PubMed] [Google Scholar]
- 12.Easter C. L., Sobecky P. A., Helinski D. R. (1997) J. Bacteriol. 179, 6472–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y., Pogliano J., Helinski D. R., Konieczny I. (2002) Mol. Microbiol. 44, 971–979 [DOI] [PubMed] [Google Scholar]
- 14.De Jonge N., Buts L., Vangelooven J., Mine N., Van Melderen L., Wyns L., Loris R. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 356–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoeffler A. J., Berger J. M. (2008) Q. Rev. Biophys. 41, 41–101 [DOI] [PubMed] [Google Scholar]
- 16.Reece R. J., Maxwell A. (1991) Crit. Rev. Biochem. Mol. Biol. 26, 335–375 [DOI] [PubMed] [Google Scholar]
- 17.Kampranis S. C., Maxwell A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14416–14421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reece R. J., Maxwell A. (1991) Nucleic Acids Res. 19, 1399–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nöllmann M., Crisona N. J., Arimondo P. B. (2007) Biochimie 89, 490–499 [DOI] [PubMed] [Google Scholar]
- 20.Couturier M., Bahassi el-M., Van Melderen L. (1998) Trends Microbiol. 6, 269–275 [DOI] [PubMed] [Google Scholar]
- 21.Anantharaman V., Aravind L. (2003) Genome Biol. 4, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoden J. B., Sellick C. A., Timson D. J., Reece R. J., Holden H. M. (2005) J. Biol. Chem. 280, 36905–36911 [DOI] [PubMed] [Google Scholar]
- 23.Dao-Thi M. H., Van Melderen L., De Genst E., Buts L., Ranquin A., Wyns L., Loris R. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1132–1134 [DOI] [PubMed] [Google Scholar]
- 24.Nieba L., Nieba-Axmann S. E., Persson A., Hämäläinen M., Edebratt F., Hansson A., Lidholm J., Magnusson K., Karlsson A. F., Plückthun A. (1997) Anal. Biochem. 252, 217–228 [DOI] [PubMed] [Google Scholar]
- 25.Smith A. B., Maxwell A. (2006) Nucleic Acids Res. 34, 4667–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidelberg J. F., Eisen J. A., Nelson W. C., Clayton R. A., Gwinn M. L., Dodson R. J., Haft D. H., Hickey E. K., Peterson J. D., Umayam L., Gill S. R., Nelson K. E., Read T. D., Tettelin H., Richardson D., Ermolaeva M. D., Vamathevan J., Bass S., Qin H., Dragoi I., Sellers P., McDonald L., Utterback T., Fleishmann R. D., Nierman W. C., White O., Salzberg S. L., Smith H. O., Colwell R. R., Mekalanos J. J., Venter J. C., Fraser C. M. (2000) Nature 406, 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bocs S., Danchin A., Médigue C. (2002) BMC Bioinformatics 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khlebnikov A., Datsenko K. A., Skaug T., Wanner B. L., Keasling J. D. (2001) Microbiology 147, 3241–3247 [DOI] [PubMed] [Google Scholar]
- 29.Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., Ehrenberg M. (2003) Cell 112, 131–140 [DOI] [PubMed] [Google Scholar]
- 30.Kamada K., Hanaoka F., Burley S. K. (2003) Mol. Cell 11, 875–884 [DOI] [PubMed] [Google Scholar]
- 31.De Jonge N., Garcia-Pino A., Buts L., Haesaerts S., Charlier D., Zangger K., Wyns L., De Greve H., Loris R. (2009) Mol. Cell 35, 154–163 [DOI] [PubMed] [Google Scholar]
- 32.Schumacher M. A., Piro K. M., Xu W., Hansen S., Lewis K., Brennan R. G. (2009) Science 323, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown B. L., Grigoriu S., Kim Y., Arruda J. M., Davenport A., Wood T. K., Peti W., Page R. (2009) PLoS Pathog. 5, e1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernard P., Couturier M. (1992) J. Mol. Biol. 226, 735–745 [DOI] [PubMed] [Google Scholar]
- 35.Kampranis S. C., Bates A. D., Maxwell A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loris R., Dao-Thi M. H., Bahassi E. M., Van Melderen L., Poortmans F., Liddington R., Couturier M., Wyns L. (1999) J. Mol. Biol. 285, 1667–1677 [DOI] [PubMed] [Google Scholar]
- 37.Dao-Thi M. H., Van Melderen L., De Genst E., Afif H., Buts L., Wyns L., Loris R. (2005) J. Mol. Biol. 348, 1091–1102 [DOI] [PubMed] [Google Scholar]
- 38.Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 4772–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 4767–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard P., Kézdy K. E., Van Melderen L., Steyaert J., Wyns L., Pato M. L., Higgins P. N., Couturier M. (1993) J. Mol. Biol. 234, 534–541 [DOI] [PubMed] [Google Scholar]
- 41.De Jonge N., Hohlweg W., Garcia-Pino A., Respondek M., Buts L., Haesaerts S., Lah J., Zangger K., Loris R. (2010) J. Biol. Chem. 285, 5606–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madl T., Van Melderen L., Mine N., Respondek M., Oberer M., Keller W., Khatai L., Zangger K. (2006) J. Mol. Biol. 364, 170–185 [DOI] [PubMed] [Google Scholar]
- 43.Oberer M., Zangger K., Gruber K., Keller W. (2007) Protein Sci. 16, 1676–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia-Pino A., Christensen-Dalsgaard M., Wyns L., Yarmolinsky M., Magnuson R. D., Gerdes K., Loris R. (2008) J. Biol. Chem. 283, 30821–30827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalton K. M., Crosson S. (2010) Biochemistry 49, 2205–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hallez R., Geeraerts D., Sterckx Y., Mine N., Loris R., Van Melderen L. (2010) Mol. Microbiol. 76, 719–732 [DOI] [PubMed] [Google Scholar]
- 47.Yamaichi Y., Fogel M. A., Waldor M. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horowitz D. S., Wang J. C. (1987) J. Biol. Chem. 262, 5339–5344 [PubMed] [Google Scholar]
- 49.Baird C. L., Harkins T. T., Morris S. K., Lindsley J. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13685–13690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nöllmann M., Stone M. D., Bryant Z., Gore J., Crisona N. J., Hong S. C., Mitelheiser S., Maxwell A., Bustamante C., Cozzarelli N. R. (2007) Nat. Struct. Mol. Biol. 14, 264–271 [DOI] [PubMed] [Google Scholar]
- 51.Kampranis S. C., Howells A. J., Maxwell A. (1999) J. Mol. Biol. 293, 733–744 [DOI] [PubMed] [Google Scholar]
- 52.Morais Cabral J. H., Jackson A. P., Smith C. V., Shikotra N., Maxwell A., Liddington R. C. (1997) Nature 388, 903–906 [DOI] [PubMed] [Google Scholar]
- 53.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lessl M., Balzer D., Lurz R., Waters V. L., Guiney D. G., Lanka E. (1992) J. Bacteriol. 174, 2493–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simic M., De Jonge N., Loris R., Vesnaver G., Lah J. (2009) J. Biol. Chem. 284, 20002–20010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.