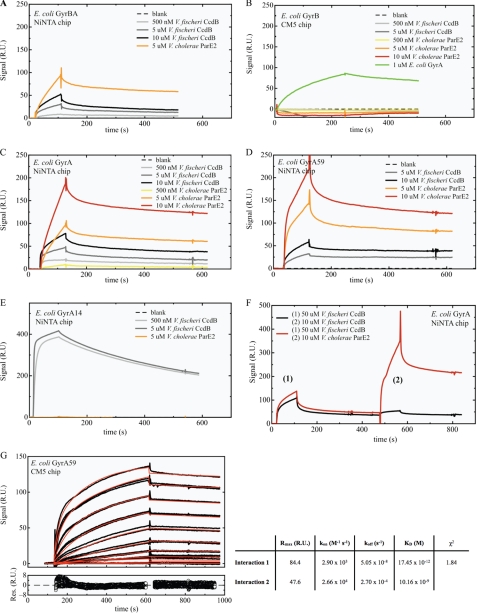
FIGURE 5.
SPR measurements of interactions between V. cholerae ParE2 or V. fischeri CcdB and gyrase. GyrBA (∼200 relative units (RU)) (A), GyrA (∼150 RU) (C), GyrA59 (∼600 RU) (D), and GyrA14 (∼400 RU) (E) were non-covalently coupled to a Ni-NTA chip, whereas GyrB (∼1800 RU) (B) was covalently coupled to a CM5 chip. The analytes were injected over the immobilized ligands at the indicated concentrations, expressed in terms of monomer. F, V. fischeri CcdB was injected at a concentration of 50 μm to saturate the CcdB binding site on GyrA. After a dissociation time of 200 s, ParE2 was injected at a concentration of 10 μm. A control experiment with a second CcdB (10 μm) injection showed that all CcdB binding sites are saturated after the first CcdB injection. G, shown is kinetic analysis of the interaction between V. cholerae ParE2 and E. coli GyrA59. The top graph displays the sensorgrams at different ParE2 concentrations (0 nm, 3.9 nm, 7.8 nm, 15.6 nm, 31.25 nm, 62.5 nm, 125 nm, 250 nm, 500 nm, 1 μm, 2 μm), which were collected in duplicate and are shown in black. The red lines represent the best fit of the model function (heterogeneous ligand model) to the experimental curves. The residuals of the fitting procedure are shown in the bottom graph. Model-based analyses of the sensorgrams recorded during the multicycle analysis of the ParE2-GyrA59 interaction indicates that the binding of ParE2 to GyrA59 is not monophasic and that two binding events occur in parallel. Fitting with a simple 1:1 Langmuir binding model does not result in acceptable residuals (χ2 = 22.4). The simplest model providing a reasonable fit to the data is a heterogeneous ligand model, resulting in a χ2 of 1.84. In aggregate, analysis of this data set reveals a very high affinity for the ParE2-GyrA59 interaction with a KD between ∼20 pm and 10 nm.