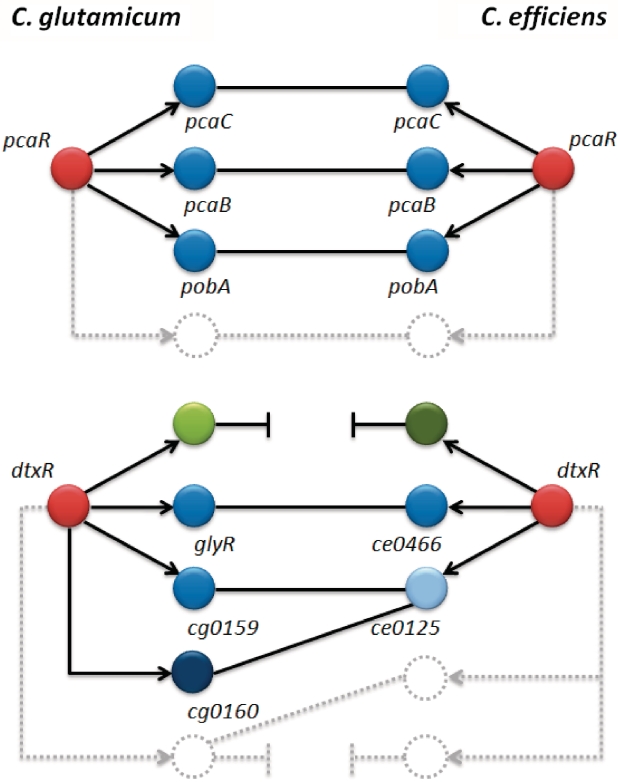
Figure 1.
Illustration of the orthology detection problem. To demonstrate this problem, we compare the regulons of the transcription factors pcaR and dtxR of Corynebacterium glutamicum (CG) and Corynebacterium efficiens (CE). The red nodes represent the respective regulators, the others their target genes. Directed edges correspond to transcriptional regulatory interactions. Undirected edges symbolize putative orthologies due to sequence-based similarity. While for pcaR all 12 target genes are conserved in both organisms, for dtxR multiple problems occur: dtxR regulates 64 genes in CG but only 27 in CE. From these target genes, only nine are clearly evolutionarily conserved, i.e. one-to-one relationship, such as glyR and ce0466. The others are either inhomologous (green nodes) or show multiple, ambiguous sequence-based similarities, i.e. one-to-many or many-to-many relationship; cg0159, cg0160 (CG), and ce0125 (CE) may serve as an example here.