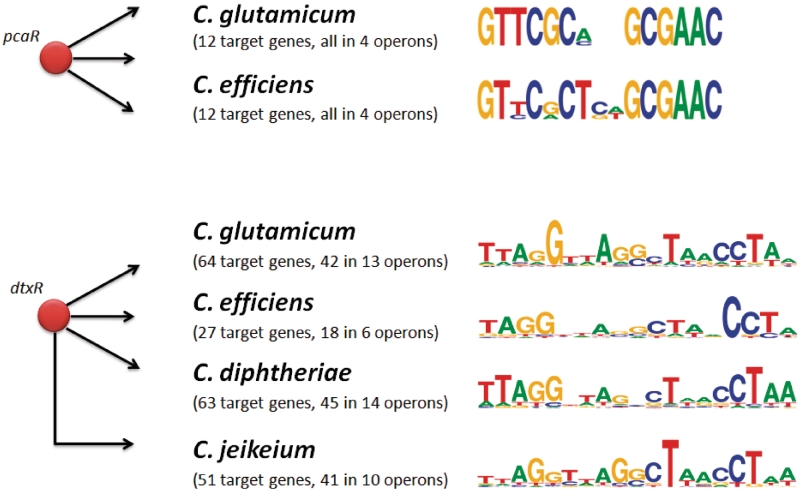
Figure 2.
Illustration of the binding-sites detection problem. Here, we demonstrate the problem when moving from one organism to another by investigating the evolutionary conservation of transcription factor binding sites. As in Figure 1, we study the transcriptional regulators pcaR in Corynebacterium glutamicum (CG) and Corynebacterium efficiens (CE) as well as the regulator dtxR in CG, CE, Corynebacterium diphtheriae (CD) and Corynebacterium jeikeium (CJ). For pcaR, all 12 target genes are conserved as are the transcription factor binding sites (TFBSs), depicted by the sequence logos (74) at the right side. It is more complicated with dtxR. The regulons are not conserved, ranging from 27 target genes in CE to 64 targets in CG. The sequence logos for DtxR are also slightly different for CG, CE, CD, and CJ.