Abstract
CAG repeats occur predominantly in the coding regions of human genes, which suggests their functional importance. In some genes, these sequences can undergo pathogenic expansions leading to neurodegenerative polyglutamine (poly-Q) diseases. The mutant transcripts containing expanded CAG repeats possibly contribute to pathogenesis in addition to the well-known pathogenic effects of mutant proteins. We have analysed two crystal forms of RNA duplexes containing CAG repeats: (GGCAGCAGCC)2. One of the structures has been determined at atomic resolution (0.95 Å) and the other at 1.9 Å. The duplexes include non-canonical A–A pairs that fit remarkably well within a regular A-helix. All the adenosines are in the anti-conformation and the only interaction within each A–A pair is a single C2-H2···N1 hydrogen bond. Both adenosines in each A–A pair are shifted towards the major groove, although to different extents; the A which is the H-bond donor stands out more (the ‘thumbs-up’ conformation). The main effect on the helix conformation is a local unwinding. The CAG repeats and the previously examined CUG structures share a similar pattern of electrostatic charge distribution in the minor groove, which could explain their affinity for the pathogenesis-related MBNL1 protein.
INTRODUCTION
Trinucleotide repeats have received special attention in biomedical research because some of them are known to undergo pathogenic expansions leading to incurable triplet repeat expansion diseases (TREDs) (1). Several TREDs are triggered by expanded triplet repeats located in UTRs of the implicated genes. These include fragile X syndrome (FXS) caused by abnormally elongated CGG repeats located in 5′-UTR of the fragile X mental retardation gene (FMR1), and myotonic dystrophy type 1 (DM1) caused by an expanded CUG repeat present in 3′-UTR of dystrophia myotonica protein kinase gene (DMPK). Most of the TREDs are triggered by expanded CAG repeat tracts that occur in protein-coding regions of specific single genes that are transcribed and translated into functionally unrelated proteins. The genetic diseases having this mutational basis include Huntington’s disease, spinal and bulbar muscular atrophy (SBMA), dentatorubral-palidoluysian atrophy (DRPLA) and several spinocerebellar ataxias (SCAs). These disorders are also known under the common name of polyglutamine (poly-Q) diseases because the CAG repeat tracts encode polyglutamine, that results in altered protein structure, shown in numerous studies to be involved in pathogenesis (2,3).
Over this decade, several authors have proposed that also mutant transcript having expanded CAG repeat may contribute to pathogenesis of polyglutamine diseases (4–9). Among the arguments used in favour of this possibility was the similarity of the RNA secondary structure formed by the CAG repeats and CUG repeats implicated in RNA-mediated pathogenesis of DM1 and SCA8 (4,10). The CAG and the CUG repeats in transcripts have been shown to interact with the same splicing regulator MBNL1 (7,11) whose sequestration by CUG tracts cause alternative splicing aberrations leading to DM1 (12) and SCA8 (13). Moreover, it was shown using repeats expressed from suitable genetic constructs that expanded non-translated CAG repeats were capable of triggering neurodegeneration in vivo in the Drosophila eye model of SCA3 (8).
The CAG repeat tracts composed of six or more repeat units are present in about 300 human genes and these sequences are strongly overrepresented in exons, which suggests their positive selection and functional importance (14). Little is known about physiological roles of CAG repeats in transcripts but their structural characterisation is more advanced. Most of the relevant structural studies have been carried out using biochemical methods on CAG repeats buried in sequence context of mRNAs of genes implicated in poly-Q disease (15–17). Short CAG repeat tracts were shown to be single-stranded, but longer repeats formed fairly stable hairpins in which alternating A–A interactions occurred between the G–C and C–G base pairs (18). Also structures formed by pure CAG repeats were compared with those formed by other triplet repeats, using both biochemical (10,20) and biophysical (19,20) methods. The CAG repeats were shown to form considerably more stable hairpins than CUG repeats of the same length (20) but the specific structural factor responsible for this difference could not be identified.
In this study, we have determined the crystal structure of an RNA duplex containing consecutive CAG repeats, based on two different crystal forms of GGCAGCAGCC. One of the structures has been determined at atomic resolution (0.95 Å). The study addresses the issue of the detailed structure as well as similarities and differences between the CAG and CUG repeat structures.
MATERIALS AND METHODS
Synthesis, purification and crystallization of CAG oligoribonucleotides
rGGCAGCAGCC oligomer was synthesized on an Applied Biosystems DNA/RNA synthesizer, using cyanoethyl phosphoramidite chemistry. Commercially available A, C and G phosphoramidites with 2'-O-tetrbutyldimethylsilyl were used for the synthesis of RNA (Glen Research, Azco, Proligo). The details of deprotection and purification of oligoribonucleotides were described previously (21). The RNA oligomer was dissolved in 100 mM KCl to the final concentration of 1 mM and annealed for 10 min at 65°C, then cooled slowly to ambient temperature within 2–3 h. Two forms of crystals were obtained, rhombohedral and trigonal, by the hanging drop/vapour diffusion method. The crystallization drops initially contained 2 μl of RNA and 2 μl of the reservoir solution. The initial volume of the reservoir solution was 500 μl. The rhombohedral crystal appeared after almost 1 year at 19°C in 25 mM MgSO4, 50 mM Tris–HCl pH 8.5 and 1.8 M (NH4)2SO4. The size of the crystal was 0.4 × 0.4 × 0.5 mm. The second form was obtained at 30°C in 10 mM MgSO4, 50 mM cacodylate-NaOH pH 6.5 and 2 M (NH4)2SO4. Crystals appeared within 2–3 weeks and then were moved to 19°C.
X-ray data collection, structure solution and refinement
X-ray diffraction data were collected: for the rhombohedral crystal on BL 14.1 beam line at the BESSY synchrotron in Berlin to the resolution of 0.95 Å and for the trigonal form on EMBL X13, DESY, Hamburg, resolution 1.9 Å. Both forms were cryoprotected by 20% glycerol (v/v) in the mother liquor. The data were integrated and scaled using the program suite DENZO/SCALEPACK (22). Although the cell parameters of the two crystal forms were similar, the space groups were different: R32:H and P32 (details in Supplementary Data). Solving the structures by molecular replacement, using PHASER (23), revealed different packing of the r(GGCAGCAGCC)2 oligomer in the same crystal cell. Early stages of the refinement were done using the program Refmac5 (24) from the CCP4 program suite (25) then refinement was carried out with PHENIX (26). Approximately 1000 reflections were set aside for the Rfree statistic (5%). The program Coot (27) was used for visualisation of electron density maps 2Fo–Fc and Fo–Fc and for manual rebuilding of the atomic model. Solvent water molecules were added by ARP/wARP (28) working in the default solvent building mode. During the refinement of the rhombohedral structure, anisotropic temperature factors was implemented and hydrogen atoms were added to the model. The last few cycles were performed using all data, including the Rfree set. The final cycles of the refinement were carried out without stereochemical restraints. The trigonal model was refined using isotropic B-factors and TLS strategy.
The helical parameters were calculated using 3DNA (29). Sequence-independent measures were used, based on vectors connecting the C1′ atoms of the paired residues, to avoid computational artefacts arising from non-canonical base pairing. Program PDB2PQR (30) was used to assign partial charges and radii to atoms of the models, according to the AMBER force field. Subsequently, the surface electrostatic potential for the RNA models was calculated with APBS (31). All pictures were drawn using PyMOL v0.99rc6 (32). The coordinates of both crystallographic models have been deposited with the Protein Data Bank (PDB). The accession codes are 3NJ6 and 3NJ7.
RESULTS
Final models
In the rhombohedral structure, the asymmetric unit contains one RNA strand (chain H). The second strand of the duplex is symmetry-related via a crystallographic 2-fold axis. In the trigonal structure, the asymmetric unit contains tree duplexes: A+B, C+D, E+F. All the duplexes stack end-to-end, forming semi-infinite columns parallel to the c cell edge. The RNA interacts with ordered water molecules and sulphate anions. The models are summarized in Table 1 and Supplementary Table S1. Crystal lattice interactions are discussed in Supplementary Data.
Table 1.
Summary of the atomic models and refinement statistics of (GGCAGCAGCC)2
| Crystal form | R32:H | P32 |
| Resolution (Å) | 0.95 | 1.9 |
| Overall mean B-factor (Å2) | 11.4 | 24.4 |
| Number of reflections: work/test | 21 748 | 15 413/821 |
| R-value (%) | 10.6 | 21.17 |
| R-free (%) | – | 24.82 |
| RNA atoms | 213 | 1278 |
| Water molecules | 86 | 170 |
| No. sulphate ions | 1 | 6 |
| R.m.s.d. in bonds (Å) | 0.016 | 0.006 |
| R.m.s.d. in angles (°) | 2.0 | 1.4 |
RNA duplex conformation and non-canonical A-A pairing
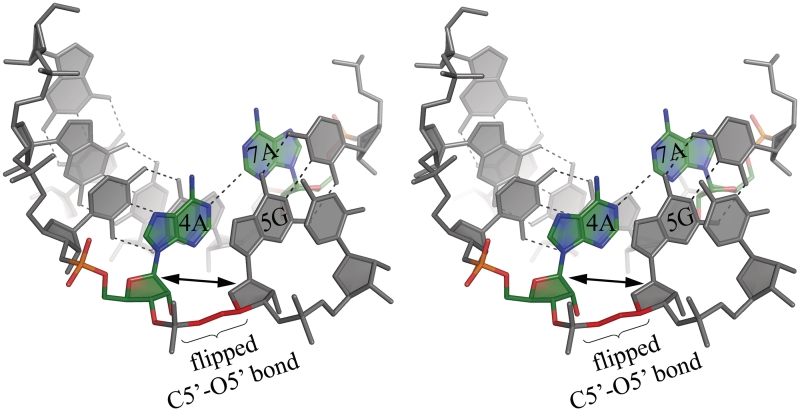
The sequence-independent helical parameters, based on inter-strand vectors between C1′ atoms, were found to be a convenient, although simplified, measure of the helix properties (Supplementary Table S2). All the duplexes are in the A-form, with the Zp values (the displacement of the phosphorus atom from the xy-plane of the ‘middle frame’ between neighbouring base-pairs) in the range 2.3–3.1 Å (33). The sugar conformation of most residues is 3′-endo, with the exception of 5G in chain A and 5G in chain E, which show the 2′-exo puckering. Values of the torsion angles fit in the typical range for the A-form (Supplementary Table S3). However, some distortions are observed, mainly for α- and γ-angles of all guanosines 5G. Instead of the common A-RNA conformation –gauche –sc, the α-angles for 5G in all seven strands are in different conformational regions: +sp for chain H, +ac for chain A, C and E, –ac for B, D and F. Only in chain B the γ-angle is in the typical +sc. It is +ap in H, –ap in A, C and E, +ac in D and F. This can be visualized as flipping of the O5′ atom due to a rotation of the O5′–C5′ bond. The effect on the conformation of the RNA strand is that the sugar rings of 5G and 4A become nearly co-planar (Figure 1). The corresponding helical twist shows unwinding of the duplex in the AG/CA steps, with helical twist values in the range 18–22°, compared to an average of 31° for other steps. Overall, the duplexes are underwound with average values of 12.5–12.9 base pairs per turn. The major groove opens up in the middle of each duplex to >20 Å (Supplementary Table S4). The inter-strand distance measured between the C1′ atoms of the paired residues is typical for A-RNA—10.7 Å, with standard deviation of 0.2 Å. It is only slightly longer (11.0 Å) for the paired adenosines.
Figure 1.
Stereo view of the A–A pair (green) and its surroundings in the (GGCAGCAGAA)2 0.95 Å resolution structure. The α- and γ-backbone torsion angles between A4 and G5 take unusual values (see text) which results in local unwinding of the helix. This can be seen by comparing the orientation of consecutive ribose rings, which appear co-planar in the two residues. This conformation of the backbone is found in all the examined A–A pairs and is associated with the adenosine less inclined towards the major groove.
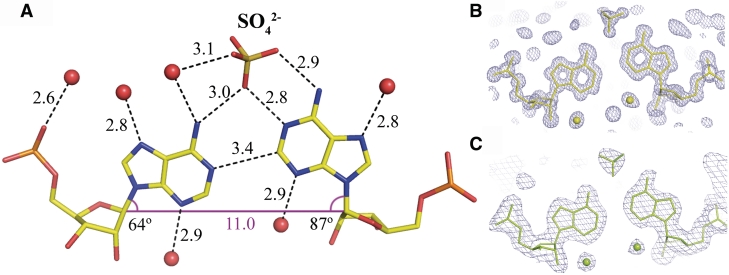
All the base pairs are well defined in the electron density and the atomic temperature factors do not show any clear patterns of variability along the RNA sequence. The C–G pairs show the Watson–Crick interactions. All the adenosines are in the anti conformation and the only interaction within each A–A pair is a single C2-H2···N1 hydrogen bond (Figure 2). In the rhombohedral, atomic resolution structure, the distance between C2 and N1 is 3.41 Å. When the H2 atom is included in the riding position its distance to N1 is 2.44 Å, which is closer by 0.3 Å than the sum of their van der Waals radii. The C, H and N atoms are almost co-linear (bond angle = 176°). Consistently with the atomic resolution data, the distances between C2 and N1 in the trigonal structure are in the range 3.1–3.4 Å. Of the two conformations that are possible in each A–A pair, the adenosine closer to the 3′-end is shifted towards the major groove, as indicated by the λ-angle (87° on average) between the bond C1′–N9 and the line between the C1′ atoms of the paired residues. The other adenosine is also upturned (λ = 64°) compared to the other residues in both structures (average λ = 55° in the range 51–58°).
Figure 2.
The A–A pair and its solvation. (A) The CH···N bond between the adenine rings is found in all the examined pairs. An associated sulphate anion is always found in the major groove but its occupancy factor is ∼0.5. The λ-angles are shown indicating the inclination of the residues towards the major groove. One adenine ring is always elevated more than the other and both are higher than average found for the C–G base pairs. The corresponding 2Fo–Fc electron density is shown for the rhombohedral (B) and a representative density for the trigonal structure (C).
Stacking interaction
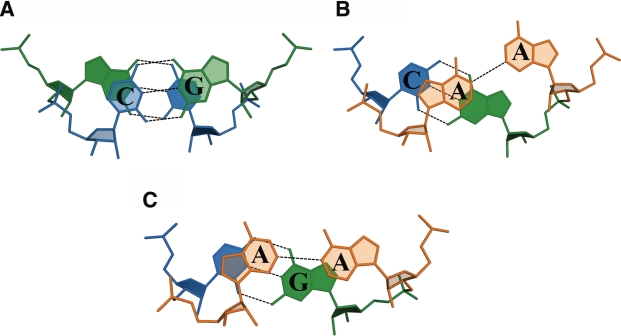
Three kinds of stacking interactions are observed in both structures: one for the GC/GC step and two for the CA/AG step, depending on the conformation of the adenosines (Figure 3). The Watson–Crick pairs show extensive overlaps typical for canonical base pairs. Reduced stacking is observed for steps involving non-canonical pairing. The more upturned adenosines stack with adjacent cytosines, on the 5′ side (average of overlap area 2.2 ± 1.1 Å2), but are far removed and do not stack with guanosines adjacent on the 3′-side. The less upturned adenosines stack to a certain degree with both their adjacent residues (0.8 ± 0.1 Å2 with C and 0.3 ± 0.3 Å2 with G).
Figure 3.
Stacking interactions in the GC/GC step (A) and two kinds of CA/AG (B and C) depending on the conformation of the A–A pair.
Hydration, ions and intermolecular interactions
The hydration of the A–A pairs forms a pattern similar in all the duplexes in the two crystal forms. In the minor groove, there is a single water molecule associated with each adenosine, bridging N3 with the O2′ atom of the ribose ring. In the major groove, there is usually a water molecule associated with N7. A sulphate ion is wedged between the paired adenine rings. One oxygen atom (O1) of the sulphate ion interacts simultaneously with N6 of the less upturned (towards the major groove) adenosine and with N1 of the other base. Another sulphate oxygen (O2) interacts with N6 of the same base (Figure 2). The interaction with the sulphate can be described as a merging of the anion binding sites described as ADE_WC_H and ADE_WC (34). The occupancy factor of the sulphate ion is ∼0.5. In its absence, a water molecule occupies the position of O1.
The hydration of C–G pairs also shows regularity, especially in the high-resolution structure. In the major groove, guanosines interact with two or three water molecules. There are always two water molecules H-bonded with the O6 and N7 atoms. The third is associated with the phosphate group. There are one or two water molecules associated with the cytosines in the major groove. One always interacts with the exo-amino group and the other is bound to the phosphate. In the minor groove, the guanosines have the capacity to interact with two water molecules: one at the exo-amino group, the other between the N3 atom and the ribose ring. However, in two cases, one of the water molecule is displaced by H-bonds formed with the oxygen atom of a symmetry-related residue. Cytosines each have one water molecule in the minor groove, H-bonded to the O2 atom.
In the trigonal lattice, there are six ribose–ribose intermolecular interactions, each consisting of four CH···O hydrogen bonds: two C1′-H1′···O2′ and two C4′-H4′···O4′. Every 3C and every 8G is involved, on each strand. The paired sugars are either between two cytidines (two pairs), two guanosines (two pairs) or between a cytidine and a guanosine (two pairs). Similarly, in the rhombohedral structure, two symmetry-related pairs are observed of 3C with 8G (Supplementary Figure S2). The average donor-acceptor distance is 3.4 Å with the standard deviation 0.1 Å.
Electrostatic potential and surface features
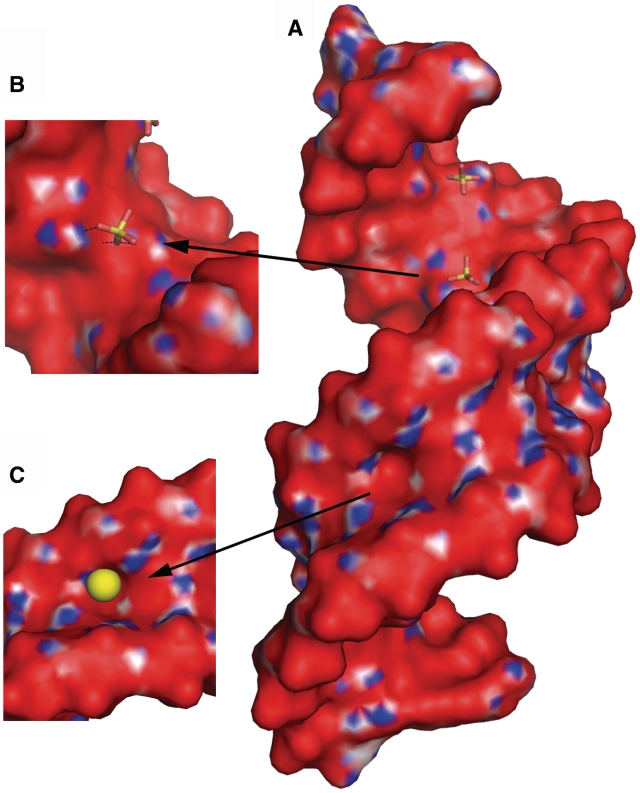
The calculated electrostatic potential shows a similar surface charge distribution for all duplexes. In the minor groove, the bands of positive and negative charge are arranged alternately along the direction of the helix axis (Figure 4). The positive bands are formed by the sugar rings and the exo-amino groups of G. The major groove is predominantly electronegative with patches of positive potential at the stacked A and C residues. The hydrogen atoms of the amino groups of each A–A pair form a stripe of electropositive potential across the major groove. These are the binding places of the sulphate ions. Adjacent cytosines generate additional positive patches on either side of the A–A pair, while the adjacent guanosines form electronegative niches in the surface of the major groove. The large shift between A and G, leading to their unstacking, results in characteristic surface features in both grooves: an indentation in the minor groove due to the protruding adenosine and a corresponding niche in the major groove at the adjacent (on the 3′-side) guanosine.
Figure 4.
The electrostatic potential surface for (A) the rhombohedral structure, showing two consecutive duplexes. Red is negative, blue is positive. Sulphate anions (sticks) are shown interacting in the major groove (detailed in B). A distinct cavity in the minor groove is filled by a water molecule (yellow sphere in C).
DISCUSSION
This work is part of the project to determine high-resolution crystal structures of all four CNG repeats, in order to identify their common and distinguishing features, which can then be interpreted in terms of their function. The atomic model interpreted in the context of the known physiochemical properties, such as the ligand affinity, surface features, electrostatic profile or hydrogen-bonding network and hydration can be used in ligand design.
One distinguishing feature of the presented structures is the A–A wobble, which to our knowledge has not been observed before. One other example of A–A pairing is found within the ribosome model (pdb code 1FFK) (35), in which both residues are in the anti conformation and interact with their Watson–Crick edges within an irregular double-stranded structure. However, the details of their interactions are different: the N6 amino group is H-bonded with N1, the two adenine rings are far from co-planar and it is doubtful if there is any significant interaction between N1 and C2 (distance 3.7 Å). The C1′–C1′ distance in the ribosomal structure is 12.4 Å—a consequence of the large size of the two purines interacting vis-a-vis. In contrast, the A–A pairs presented here, embedded in CAG repeats, fit remarkably well within a regular A-helix. Although accommodation of the interacting purine rings seems to be sterically demanding, the inter-strand C1′–C1′ distances for the adenosine residues are only slightly larger than average. This ability to conform to the helical form is worth noting in view of the fact that in the literature the CNG double stranded forms are often referred to as ‘containing internal loops’. In terms of the 3D structure, the main consequence of an A–A pair seems to be a local unwinding of the duplex. For the adenosine that is inclined towards the minor groove, the α-torsion angle of the following G takes positive or high negative values, and γ takes high values (Figure 1, Supplementary Table S3). Consequently, the two sugar rings on either side of the phosphate group are almost co-planar, as opposed to the typical case, in which the successive sugar rings follow the helical twist. The values of the twist associated with A–A and the following pair are small (in the range 18–22°) compared with the average value of 31–32° for other steps in the duplexes.
The only interaction between the paired adenosine residues is the weak C2-H2···N1 bond. Carbon is a poorer donor than nitrogen or oxygen and the later two clearly dominate in the H-bonding interactions in biological molecules. The energy of C-H···X bonds is estimated to be ∼1 kcal/mol or less, with the C-H···N bonds being weaker, less frequent and poorly studied compared to C-H···O. The energies, although small, are not negligible and correspond to measurable effects on the thermal stability of the duplex. Remarkably, in a thermodynamic study of related RNA sequences (19), bromination of one adenine, which is expected to force the adenosine residue into the syn conformation, results in a destabilisation of the helix, as indicated by an increase of the free energy of duplex formation by ∼0.7 kcal/mol, with a decrease of the melting temperature by a corresponding 4°C. The effect seems to be additive when more A–A pairs are modified.
In order to assess the biological significance, one needs to consider the present structure in the wider context of CNG repeats. Pathogenesis involving expanded runs of CAG repeats is well known to occur at the protein level. However, the role of the transcripts should also be considered. In one type of spinocerebellar ataxia the abnormal CAG run is found only in the UTR, the 5′-UTR promoter (36). In binding studies of MBNL1 protein, CAG repeats show similar affinity as CUG runs, both in vitro and in vivo (7,11,37). Detailed and well-parameterized structural model is necessary to explain the physiochemical properties of CAG structures and to use them as targets for ligands to block the translation of poly-Q mutant proteins. When comparing the present structure of the CAG repeats and the previously described CUG repeats (38) one can observe both similarities and differences (Table 2 and Supplementary Figures S3–5). In terms of the overall helical twist the CAG-containing duplexes are underwound (12.5–12.9 bp/turn), while CUG structures are more typical (11 bp/turn). The major groove in the CAG helices is wide and shallow, while in CUG it is narrow and deep. Each A–A and U–U pair can assume two alternative relative positions, depending on which base is the H-bond donor or acceptor (as it happens, the acceptor in both types of repeat is more inclined towards the minor groove). The four CAG-containing duplexes in the two crystal structure are closely superposable and they all show the same order of A–A paring conformations. The first adenosine, from the 5′-end, always acts as the H-bond acceptor within the A–A pair and the second A is in the ‘thumbs-up’ conformation, pointing towards the major groove, and acts as the H-bond donor. This is not a consequence of crystal symmetry, at least in the case of the trigonal structure, in which the three duplexes are crystallographically independent. The observed structures all correspond to one of three theoretically foreseeable arrangements of two consecutive A–A pairs (the other, unobserved, arrangements of the two adenosines within a strand would have alternating but reversed inclinations or similar inclinations). This can be contrasted with the structure of (CUG)n duplexes which show an apparently random order of two possible U–U pairing conformations within the CUG repeats (38).
Table 2.
Comparison of characteristic features of double helical CAG and CUG (38) repeats
| Feature | CAG | CUG |
| Helix form | A | A |
| Helical twista (°) | 28.5 ± 5.7 | 33.6 ± 4.1 |
| Major groove widthb (Å) | 23.8 ± 0.2 | 12.7 ± 2.3 |
| Minor groove widthb (Å) | 15.3 ± 0.1 | 15.7 ± 0.4 |
| Average C1′–C1′ distance for N–Nc (and for the other pairs) (Å) | 11.0 (10.7) | 10.4 (10.5) |
| Local effect of N–N on helicity | unwinding | not observed |
| N–N pairing interaction | C2-H2···N1 hydrogen bond | N3-H3···O4 hydrogen bond |
| Manner of accommodating N–N (according to direction of the glycosidic bond, λ) | One A turned towards major groove (‘thumbs-up’) | One U inclined towards minor groove |
| Effect of N–N conformation on neighbouring N–N | Cooperativity: A–A pairs in consecutive repeats have alternative conformations | No effect: each U–U takes one of two possible conformation independently |
| Electrostatic profile | Alternating stripes of positive and negative potential due to C–G pairs | Alternating stripes of positive and negative potential due to C–G pairs |
| Observed ligand affinity of N–N | Sulphate binding in major groove | Sulphate or glycerol through-water binding in major groove |
| Exposed functional groups of N–N | ||
| Major groove | First A N6 amino Second A N1 imino, N6 amino | First U O4 carbonyl |
| Minor groove | First A N3 imino Second A N3 imino | First U O2 carbonyl Second U O2 carbonyl, N3 amino |
One clear similarity between CAG- and CUG-duplexes is the pattern of stripes of alternating positive and negative electrostatic potential in the minor groove. The structural basis of the pattern is similar in both types of repeats except that the negative potential in the CAG structure is due to the imine groups of the adenosine residues, while in the CUG structure this is due to the carbonyl oxygen atoms of the uridines. This could explain some features of the repeat tracts, such as the reported affinity of the MBNL1 protein for both CUG and CAG (38). In the case of CCG repeats (whose structure is still unknown), which are also recognized by the MBNL1 protein, the C–C pairs should also contribute electronegative potential in the minor groove, due to their carbonyl groups. In contrast, the CGG-containing duplexes, which do not interact directly with the MBNL1 protein (40), are expected to present in the minor groove at least one prominent amine group. In the major groove, on the other hand, the A–A pair shows an affinity for sulphate ions. This is apparently due to the exposed Watson–Crick edge of the adenosine in the ‘thumbs-up’ conformation. The binding of the sulphate is a consequence of high concentration of the anions in the crystallization medium and is unlikely to take place to a significant degree in the cell, but the interaction in the crystals can be taken as an indication of an affinity of the exposed Watson–Crick edge of the adenine for negatively charged bidentate ligands.
ACCESSION NUMBERS
The accession numbers are PDB 3NJ6, 3NJ7.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Higher Education (Poland, N-N301-0171634, PBZ-MNiSW-07/I/2007, PBZ-MNiI-2/1/2005, PBZ-KBN-124/P05/2004); the European Community – Research Infrastructure Action under the FP6 ‘Structuring the European Research Area’ Programme (through the ‘Integrated Infrastructure Initiative’ Integrating Activity on Synchrotron and Free Electron Laser Science – Contract R II 3-CT-2004-506008); Fellowship of the Foundation for Polish Science (to R.K.). Funding for open access charge: Research Grant.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J. Biol. Chem. 2009;284:7425–7429. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galvão R, Mendes-Soares L, Câmara J, Jaco I, Carmo-Fonseca M. Triplet repeats, RNA secondary structure and toxic gain-of-function models for pathogenesis. Brain Res. Bull. 2001;56:191–201. doi: 10.1016/s0361-9230(01)00651-7. [DOI] [PubMed] [Google Scholar]
- 5.Broude NE, Cantor CR. Neurological diseases and RNA-directed gene regulation: prospects for new diagnostics and therapy. Expert Rev. Mol. Diagn. 2003;3:269–274. doi: 10.1586/14737159.3.3.269. [DOI] [PubMed] [Google Scholar]
- 6.Jasinska A, Michlewski G, de Mezer M, Sobczak K, Kozlowski P, Napierala M, Krzyzosiak WJ. Structures of trinucleotide repeats in human transcripts and their functional implications. Nucleic Acids Res. 2003;31:5463–5468. doi: 10.1093/nar/gkg767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 8.Li L-B, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birman S. Neurodegeneration: RNA turns number one suspect in polyglutamine diseases. Curr. Biol. 2008;18:R659–R661. doi: 10.1016/j.cub.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LPW. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michlewski G, Krzyzosiak WJ. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Sobczak K, Krzyzosiak WJ. Imperfect CAG repeats form diverse structures in SCA1 transcripts. J. Biol. Chem. 2004;279:41563–41572. doi: 10.1074/jbc.M405130200. [DOI] [PubMed] [Google Scholar]
- 17.Sobczak K, Krzyzosiak WJ. CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. J. Biol. Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
- 18.Krzyzosiak WJ, Sobczak K, Napierala M. In: Genetic Instabilities and Neurological Diseases. Wells RD, editor. Elsevier; 2006. pp. 705–716. [Google Scholar]
- 19.Broda M, Kierzek E, Gdaniec Z, Kulinski T, Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–10882. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
- 20.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–325. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational P. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr. D Biol. Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 29.Olson WK, Bansal M, Burley SK, Dickerson RE, Gerstein M, Harvey SC, Heinemann U, Lu XJ, Neidle S, Shakked Z, et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 30.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLano WL. The PyMOL Molecular Graphics System. CA, USA: DeLano Scientific, Palo Alto; 2002. [Google Scholar]
- 33.Lu X-J, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auffinger P, Bielecki L, Westhof E. Anion binding to nucleic acids. Structure. 2004;12:379–388. doi: 10.1016/j.str.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 36.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 38.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the ‘stretched U-U wobble’: implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bloomfield VA, Crothers DM, Tinoco I. Nucleic Acids: Structures, Properties, and Functions. University Science Books, Sausalito; 2000. [Google Scholar]
- 40.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.